Three New Human Genes: De Novo Genes
Entirely novel human-specific protein-coding genes originating from ancestrally noncoding sequences are reported by two geneticists at the University of Dublin (1). Analyzing available data, they identified genes that are expressed in the human species but not in chimps. They then looked for simiar sequences in other primates, finding three. The chimp and macaque (unexpressed) sequences are nearly identical to the human one, but are interrupted by frameshifting insertions and stop codons. |
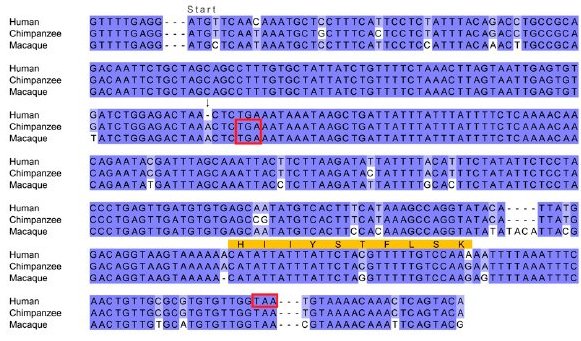 |
| "Multiple sequence alignment of the gene sequence of the human gene CLLU1 and similar nucleotide sequences from the syntenic location in chimp and macaque. The start codon is located immediately following the first alignment gap, which was inserted for clarity. Stop codons are indicated by red boxes. The sequenced peptide identified from this locus is indicated in orange. The critical mutation that allows the production of a protein is the deletion of an A nucleotide, which is present in both chimp and macaque (indicated by an arrow). This causes a frameshift in human that results in a much longer ORF capable of producing a 121-amino acids-long protein. Both the chimp and macaque sequences have a stop codon after only 42 potential codons." © Genome Research 2009 |
CLLU1 is also disabled by a matching point insertion in the gorilla and gibbon, but not orangutan, genomes. The geneticists reason, If the ancestral primate sequence was coding, then we would need to infer that an identical 1-bp insertion occurred in four lineages independently, whereas if we infer the presence of the disabler in the ancestral sequence, then we must infer two independent 1-bp deletions. The inference that the ancestral sequence was noncoding is a more parsimonious explanation of the data, even without considering that the parallel insertion of a specific base into an identical location is probably less likely than the parallel deletion of one base. ...We hypothesize that these genes have originated de novo in the human lineage, since the divergence with chimp from ancestrally noncoding sequence.
| Consider a protein of only 120 amino acids in length. Assume that the protein needs ~25%, or 30, of its amino acids exactly right. There are 20 amino acids, so only 1 out of 20 amino acids can occupy each of those 30 positions. The chance that 30 random amino acids will match this sequence in one trial can be estimated as
(1/20)^30 = ~10^-40 Assume that the remaining 90 amino acids in this sequence may vary widely, such that any 10 of life's 20 amino acids will do. The chance that 90 random amino acids will satisfy these criteria in one trial is approximately (10/20)^90 = ~10^-28 Combining these assumptions, the chance that a given sequence of 120 random amino acids will constitute a working version of this protein is on the order of 10^(-40-28) = 10^-68 (This method copies Chandra Wickramasinghe's in The Scientific Legacy of Fred Hoyle, reviewed, 2005.) |
The claim that these genes, "originated de novo in the human lineage," is baffling. Sequences virtually identical to them already exist in species considered ancestral to humans, and even in mice. The genes "were activated" is a more accurate description.
And the geneticists' use of math is interesting: At one location, two deletions are more likely than four identical insertions, so the ancestral sequence must have been noncoding? Why not ask how likely it is that a sequence of 121 codons — apparently unaffected by natural selection — happens to encode a given working protein? By our analysis (see box at right), it is forbiddingly unlikely, even if this relatively small protein could vary widely. It's the monkeys writing Shakespeare again.
The darwinian explanation of this phenomenon is unclear to us. But the discovery neatly aligns with cosmic ancestry, which predicts:
![]() ...Genes precede the phenotypic expression of themselves (2).
...Genes precede the phenotypic expression of themselves (2).
![]() If a new genetic program arrives as cosmic ancestry predicts, intervening (ancestral) species should possess either nearly identical versions of it ...or nothing similar... (3).
If a new genetic program arrives as cosmic ancestry predicts, intervening (ancestral) species should possess either nearly identical versions of it ...or nothing similar... (3).
![]() ...At least some of the silent DNA is for future use (4).
...At least some of the silent DNA is for future use (4).
![]() Point mutations and other simple mechanisms can switch existing programs off and on (5).
Point mutations and other simple mechanisms can switch existing programs off and on (5).
![]() ...This process would ...depend on sophisticated software management that can recognize an installed program (6).
...This process would ...depend on sophisticated software management that can recognize an installed program (6).
![]() New genetic programs will be continually offered for testing (7).
New genetic programs will be continually offered for testing (7).
What'sNEW since 2009
![]() 07 Jun 2024: De novo genes by reverse transcription of a "rolling circle" of RNA?
07 Jun 2024: De novo genes by reverse transcription of a "rolling circle" of RNA?
![]() "De novo genes: from non-genic to genic," by Li Zhao, Nature Reviews Genetics, Feb 2024.
...more recent research has shown that a small sub-set of these young, lineage-restricted genes arises de novo from ancestrally non-genic sequences.
"De novo genes: from non-genic to genic," by Li Zhao, Nature Reviews Genetics, Feb 2024.
...more recent research has shown that a small sub-set of these young, lineage-restricted genes arises de novo from ancestrally non-genic sequences.
![]() "Generation of de novo miRNAs from template switching during DNA replication," by Heli A. M. Mönttinen et al., PNAS, 29 Nov 2023.
...template switching ...allows for near-instant rewiring of genetic information.... and commentary:
"Generation of de novo miRNAs from template switching during DNA replication," by Heli A. M. Mönttinen et al., PNAS, 29 Nov 2023.
...template switching ...allows for near-instant rewiring of genetic information.... and commentary:
![]() "New genes found that can arise 'from nothing'," University of Helsinki via PhysOrg, 08 Dec 2023.
"New genes found that can arise 'from nothing'," University of Helsinki via PhysOrg, 08 Dec 2023.
![]() 03 May 2023: Can de novo genes emerge via a preferred trajectory?
03 May 2023: Can de novo genes emerge via a preferred trajectory?
![]() 22 Mar 2023: ...de novo genes that influence the growth of the human brain.
22 Mar 2023: ...de novo genes that influence the growth of the human brain.
![]() "De novo genes with an lncRNA origin encode unique human brain developmental functionality," by N.A. An, J. Zhang, F. Mo et al., doi:10.1038/s41559-022-01925-6, Nat Ecol Evol, 02 Jan 2023.
It is ...difficult to understand the process by which a de novo gene acquires its biological function.
"De novo genes with an lncRNA origin encode unique human brain developmental functionality," by N.A. An, J. Zhang, F. Mo et al., doi:10.1038/s41559-022-01925-6, Nat Ecol Evol, 02 Jan 2023.
It is ...difficult to understand the process by which a de novo gene acquires its biological function.
![]() De novo birth of functional microproteins in the human lineage by Nikolaos Vakirlis et al., doi:10.1016/j.celrep.2022.111808, Cell Reports, 20 Dec 2022.
...the intriguing, and still largely mysterious phenomenon of de novo gene birth....
De novo birth of functional microproteins in the human lineage by Nikolaos Vakirlis et al., doi:10.1016/j.celrep.2022.111808, Cell Reports, 20 Dec 2022.
...the intriguing, and still largely mysterious phenomenon of de novo gene birth....
![]() A putative de novo evolved gene required for spermatid chromatin condensation in Drosophila melanogaster by Emily L. Rivard, Andrew G. Ludwig, Prajal H. Patel et al., doi:10.1371/journal.pgen.1009787, PLoS Genet., 03 Sep 2021.
A putative de novo evolved gene required for spermatid chromatin condensation in Drosophila melanogaster by Emily L. Rivard, Andrew G. Ludwig, Prajal H. Patel et al., doi:10.1371/journal.pgen.1009787, PLoS Genet., 03 Sep 2021.
![]() 14 Aug 2021: "...land plants ...acquiring de novo genes"
14 Aug 2021: "...land plants ...acquiring de novo genes"
![]() 10 Dec 2020: Mutualisms between fungi and plants ...have evolved repeatedly and independently many times.
10 Dec 2020: Mutualisms between fungi and plants ...have evolved repeatedly and independently many times.
![]() Stochastic Gain and Loss of Novel Transcribed Open Reading Frames in the Human Lineage by Daniel Dowling, Jonathan F Schmitz and Erich Bornberg-Bauer, doi:10.1093/gbe/evaa194, Genome Biology and Evolution, Nov 2020. Although de novo emerged proteins have been identified in numerous organism, how they evolve and transition from chance transcriptional events to fully fledged proteins is little understood. Here we show that over the short time scale of primate evolution, the sequence properties ...of expressed human open reading frames change little.
Stochastic Gain and Loss of Novel Transcribed Open Reading Frames in the Human Lineage by Daniel Dowling, Jonathan F Schmitz and Erich Bornberg-Bauer, doi:10.1093/gbe/evaa194, Genome Biology and Evolution, Nov 2020. Although de novo emerged proteins have been identified in numerous organism, how they evolve and transition from chance transcriptional events to fully fledged proteins is little understood. Here we show that over the short time scale of primate evolution, the sequence properties ...of expressed human open reading frames change little.
![]() 02 Nov 2020: "...we call 'invention' those gene families for which predecessors (homologs outside eukaryotes) cannot be identified."
02 Nov 2020: "...we call 'invention' those gene families for which predecessors (homologs outside eukaryotes) cannot be identified."
![]() 20 Feb 2020: Orphan genes do not often emerge from the divergence of predecessor genes.
20 Feb 2020: Orphan genes do not often emerge from the divergence of predecessor genes.
![]() 19 Oct 2019: Genes-in-waiting is one way to describe de novo genes.
19 Oct 2019: Genes-in-waiting is one way to describe de novo genes.
![]() "A de novo evolved gene in the house mouse regulates female pregnancy cycles," by Chen Xie et al., doi:10.7554/eLife.44392, eLife, 2 Aug 2019. ... our findings support the hypothesis that a de novo evolved gene can directly adopt a function without much sequence adaptation.
"A de novo evolved gene in the house mouse regulates female pregnancy cycles," by Chen Xie et al., doi:10.7554/eLife.44392, eLife, 2 Aug 2019. ... our findings support the hypothesis that a de novo evolved gene can directly adopt a function without much sequence adaptation.
![]() 27 May 2019: de novo gene birth
27 May 2019: de novo gene birth
![]() De Novo, Divergence, and Mixed Origin Contribute to the Emergence of Orphan Genes in Pristionchus Nematodes by Neel Prabh and Christian Rödelsperger, doi:10.1534/g3.119.400326, G3, 05 May 2019. ...the majority of eukaryotic genomes contain large numbers of orphan genes lacking homologs in other taxa.
De Novo, Divergence, and Mixed Origin Contribute to the Emergence of Orphan Genes in Pristionchus Nematodes by Neel Prabh and Christian Rödelsperger, doi:10.1534/g3.119.400326, G3, 05 May 2019. ...the majority of eukaryotic genomes contain large numbers of orphan genes lacking homologs in other taxa.
![]() 01 May 2019: ...the Medusavirus and the origin of eukaryotic life (lots of de novo genes.)
01 May 2019: ...the Medusavirus and the origin of eukaryotic life (lots of de novo genes.)
![]() Genes that evolve from scratch expand protein diversity, Newswise, 11 Mar 2019. The research shows that random, noncoding sections of DNA can quickly evolve to produce new proteins. (evolve to?)
Genes that evolve from scratch expand protein diversity, Newswise, 11 Mar 2019. The research shows that random, noncoding sections of DNA can quickly evolve to produce new proteins. (evolve to?)
![]() Molecular mechanism and history of non-sense to sense evolution of antifreeze glycoprotein gene in northern gadids by Xuan Zhuang et al., doi:10.1073/pnas.1817138116, PNAS, online 14 Feb 2019. We experimentally proved that all genic components of the extant gadid AFGP originated from entirely nongenic DNA. ...also... a fully formed ORF existed before the regulatory element to activate transcription was acquired.
Molecular mechanism and history of non-sense to sense evolution of antifreeze glycoprotein gene in northern gadids by Xuan Zhuang et al., doi:10.1073/pnas.1817138116, PNAS, online 14 Feb 2019. We experimentally proved that all genic components of the extant gadid AFGP originated from entirely nongenic DNA. ...also... a fully formed ORF existed before the regulatory element to activate transcription was acquired.
![]() 21 Sep 2018: De novo genes continue to confound standard darwinism.
21 Sep 2018: De novo genes continue to confound standard darwinism.
![]() 13 Jun 2018: Pandora viruses have more than a thousand genes. At least two-thirds are completely novel.
13 Jun 2018: Pandora viruses have more than a thousand genes. At least two-thirds are completely novel.
![]() 15 May 2018: ...many genes typically associated with metazoan functions actually pre-date animals themselves....
15 May 2018: ...many genes typically associated with metazoan functions actually pre-date animals themselves....
![]() "Orphans and new gene origination, a structural and evolutionary perspective," by Sara Light, Walter Basile and Arne Elofsson, doi:10.1016/j.sbi.2014.05.006, v 26 Current Opinion in Structural Biology, online 13 Jun 2014. ...at some time in history the first protein coding sequence within a protein family must have been created from non-coding genetic material [if evolution is strictly darwinian.]
"Orphans and new gene origination, a structural and evolutionary perspective," by Sara Light, Walter Basile and Arne Elofsson, doi:10.1016/j.sbi.2014.05.006, v 26 Current Opinion in Structural Biology, online 13 Jun 2014. ...at some time in history the first protein coding sequence within a protein family must have been created from non-coding genetic material [if evolution is strictly darwinian.]
![]() The Evolution of Venom by Co-option of Single-Copy Genes by Ellen O. Martinson, Mrinalini, et al., doi:10.1016/j.cub.2017.05.032, Current Biology, 10 Jul 2017. We propose that co-option of single-copy genes may be a common but relatively understudied mechanism of evolution for new gene functions, particularly under conditions of rapid evolutionary change.
The Evolution of Venom by Co-option of Single-Copy Genes by Ellen O. Martinson, Mrinalini, et al., doi:10.1016/j.cub.2017.05.032, Current Biology, 10 Jul 2017. We propose that co-option of single-copy genes may be a common but relatively understudied mechanism of evolution for new gene functions, particularly under conditions of rapid evolutionary change.
![]() 22 May 2017: It has become clear that protein-coding genes can originate de novo from non-coding sequences.
22 May 2017: It has become clear that protein-coding genes can originate de novo from non-coding sequences.
![]() Aoife McLysaght and Daniele Guerzoni, "New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation" [pdf], doi:10.1098/rstb.2014.0332, Philosophical Transactions B, 2015. ...It has now become clear that de novo origin of protein-coding genes from non-coding DNA is a consistent feature of eukaryotic genomes....
Aoife McLysaght and Daniele Guerzoni, "New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation" [pdf], doi:10.1098/rstb.2014.0332, Philosophical Transactions B, 2015. ...It has now become clear that de novo origin of protein-coding genes from non-coding DNA is a consistent feature of eukaryotic genomes....
![]() 21 Aug 2016: Can antagonistic evolution compose de novo genes?
21 Aug 2016: Can antagonistic evolution compose de novo genes?
![]() 4 Sep 2015: ...Thousands of transcripts ...which are likely to have originated de novo....
4 Sep 2015: ...Thousands of transcripts ...which are likely to have originated de novo.... ![]() 4 Jan 2016.
4 Jan 2016.
![]() A Surprise Source of Life's Code by Emily Singer, Quanta Magazine (+Scientific American), 18 Aug 2015. Of the 600 human-specific genes that [evolutionary biologist Mar] Albà's team found, 80 percent are entirely new, having never been identified before.
A Surprise Source of Life's Code by Emily Singer, Quanta Magazine (+Scientific American), 18 Aug 2015. Of the 600 human-specific genes that [evolutionary biologist Mar] Albà's team found, 80 percent are entirely new, having never been identified before.
![]() 8 Oct 2014: 24 hominoid-specific de novo protein-coding genes were identified.
8 Oct 2014: 24 hominoid-specific de novo protein-coding genes were identified.
![]() 24 Jan 2014: The earliest steps in de novo gene origination remain mysterious.
24 Jan 2014: The earliest steps in de novo gene origination remain mysterious.
![]() Diethard Tautz, "The Discovery of De Novo Gene Evolution" [link], doi:10.1353/pbm.2014.0006, Perspectives in Biology and Medicine, Winter 2014.
Diethard Tautz, "The Discovery of De Novo Gene Evolution" [link], doi:10.1353/pbm.2014.0006, Perspectives in Biology and Medicine, Winter 2014.
![]() Bétermier M, Bertrand P, Lopez BS, "Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?" [html], doi:10.1371/journal.pgen.1004086, 10(1): e1004086, PLoS Genet, 16 Jan 2014. "...Recent data have pointed to the intrinsic precision of NHEJ."
Bétermier M, Bertrand P, Lopez BS, "Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?" [html], doi:10.1371/journal.pgen.1004086, 10(1): e1004086, PLoS Genet, 16 Jan 2014. "...Recent data have pointed to the intrinsic precision of NHEJ."
![]() Dong-Dong Wu and Ya-Ping Zhang, "Evolution and Function of De Novo Originated Genes" [abstract], Molecular Phylogenetics and Evolution, online 27 Feb 2013. Wu and Zhang think sequences evolve as proteins, but in most cases, the nucleotides are already ordered before the first translation.
Dong-Dong Wu and Ya-Ping Zhang, "Evolution and Function of De Novo Originated Genes" [abstract], Molecular Phylogenetics and Evolution, online 27 Feb 2013. Wu and Zhang think sequences evolve as proteins, but in most cases, the nucleotides are already ordered before the first translation.
![]() 25 Jan 2013: Many of our genes have no obvious relatives or evolutionary history. So where did they come from?
25 Jan 2013: Many of our genes have no obvious relatives or evolutionary history. So where did they come from?
![]() 23 Oct 2012: Evolution by subfunctionalization
23 Oct 2012: Evolution by subfunctionalization
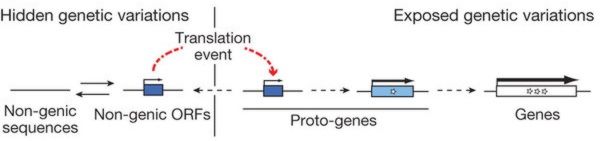
![]() A-R Carvunis et al., "Proto-genes and de novo gene birth," doi:10.1038/nature11184, Nature, online 14 Jun 2012.
A-R Carvunis et al., "Proto-genes and de novo gene birth," doi:10.1038/nature11184, Nature, online 14 Jun 2012.
|
We argue ...that the existence of orphan genes – so called because they lack homologues in other lineages – tells another story. The evolutionary origin of such genes is still unclear, even though they represent up to one-third of the genes in all genomes, including those of bacteria, archaea and phages.
– Tautz and Domazet-Loŝo, 2011 |
References
1.2.
3.
4.
5.
6.
7.