What'sNEW Nov - Dec 2020
| 07 Dec 2020 |
What'sNEW about HGT  | | |
...COVID-19 sequences may one day become one of the many retro-elements that litter the human genome.
 SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome by Liguo Zhang et al., doi:10.1101/2020.12.12.422516, bioRxiv, 13 Dec 2020. SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome by Liguo Zhang et al., doi:10.1101/2020.12.12.422516, bioRxiv, 13 Dec 2020.
 Thanks, Ted Steele. Thanks, Ted Steele.
 Viruses... has background and links. Viruses... has background and links.
...there is something seriously wrong with our understanding of the cosmos.
 ...a Cosmic Crisis Gets Worse by Natalie Wolchover, Quanta, 17 Dec 2020; re: ...a Cosmic Crisis Gets Worse by Natalie Wolchover, Quanta, 17 Dec 2020; re:
 Cosmic Distances Calibrated to 1% Precision with Gaia EDR3 Parallaxes and Hubble Space Telescope Photometry of 75 Milky Way Cepheids Confirm Tension with LambdaCDM [abstract | pdf] by Adam G. Riess et al., arXiv:2012.08534 [astro-ph.CO], 15 Dec 2020. Cosmic Distances Calibrated to 1% Precision with Gaia EDR3 Parallaxes and Hubble Space Telescope Photometry of 75 Milky Way Cepheids Confirm Tension with LambdaCDM [abstract | pdf] by Adam G. Riess et al., arXiv:2012.08534 [astro-ph.CO], 15 Dec 2020.
 The universe is expanding too fast, and that could rewrite cosmology by Stuart Clark, NewScientist, 25 Nov 2020. The universe is expanding too fast, and that could rewrite cosmology by Stuart Clark, NewScientist, 25 Nov 2020.
 The End... and The End... and
 The Beginning have related discussion and updates. The Beginning have related discussion and updates.
 Patricia Ann Straat died on October 23rd. In the 1970s, she designed, planned and conducted the Viking LR life-detection experiments for NASA, with Gil Levin. On 30 July 1976, consistent positive results backed by controls began to arrive — Life on Mars!
Patricia Ann Straat died on October 23rd. In the 1970s, she designed, planned and conducted the Viking LR life-detection experiments for NASA, with Gil Levin. On 30 July 1976, consistent positive results backed by controls began to arrive — Life on Mars!
 Patricia Ann Straat, 1936–2020, Astrobiology, 14 Dec 2020. Patricia Ann Straat, 1936–2020, Astrobiology, 14 Dec 2020.
 Patricia Ann Straat: homepage and book review. Patricia Ann Straat: homepage and book review.
 ...Viking with Dr. Patricia Ann Straat: Youtube video posted 30 Dec 2019. ...Viking with Dr. Patricia Ann Straat: Youtube video posted 30 Dec 2019.
 To Mars With Love: her book reviewed here, 23 Feb 2019. To Mars With Love: her book reviewed here, 23 Feb 2019.
 Life on Mars! discusses the LR experiments, with updates. Life on Mars! discusses the LR experiments, with updates.
Mutualisms between fungi and plants appear complex but have evolved repeatedly and independently many times. This convergent evolution is typically explained by gene loss from fungi, but at the origin of a symbiosis new genes also appear in fungi: where do these new genes come from ...? — Wang et al., 2020
Where genes come from is our question as well. Now four specialists in botany and related fields have focussed on symbiotic Amanita mushrooms and their non-symbiotic sister species. Which genes are unique to the symbiotic species? How were they acquired? The biologists identified three mechanisms of acquisition.
Mechanisms generating new gene families include de novo gene birth, horizontal gene transfer, and neofunctionalization following a duplication event. ...The origins of seven of the unique gene families are strongly supported as either de novo gene birth (two gene families), horizontal gene transfer (four), or gene duplication (one).
 Three mechanisms of gene family emergence. (A) de novo gene birth; (B) HGT; (C) gene duplication. Blue, species tree; Red, new gene families (arrow indicates origin); Black, homologous genes.
Three mechanisms of gene family emergence. (A) de novo gene birth; (B) HGT; (C) gene duplication. Blue, species tree; Red, new gene families (arrow indicates origin); Black, homologous genes.
|
De novo genes come from gene-length sequences that were formerly silent but contain few or no stop codons. They show no evidence of having been composed by darwinian trial-and-error. Genes from HGT, where they are found, often also lack such evidence. Duplicates might show evidence of re-composing by trial-and-error if the path to the new function, "neofunctionalization," is apparent. In this instance, it isn't.
The limited evidence of positive selection and the ability to differentiate the functions of the newly duplicated genes and their paralogs prevents any inference of neofunctionalization following duplication for this family.
For historical reconstructions like this, a flood of data from newly-sequenced genomes comes continually. When I ask where genes come from, I still find only genes that "seem to have come from nowhere." I welcome pointers to plausible evidence of genes that are composed from scratch by trial-and-error.
 De Novo Gene Birth, Horizontal Gene Transfer, and Gene Duplication as Sources of New Gene Families Associated with the Origin of Symbiosis in Amanita by Yen-Wen Wang, Jaqueline Hess, Jason C Slot and Anne Pringle, dio:10.1093/gbe/evaa193, Genome Biology and Evolution, Nov 2020. De Novo Gene Birth, Horizontal Gene Transfer, and Gene Duplication as Sources of New Gene Families Associated with the Origin of Symbiosis in Amanita by Yen-Wen Wang, Jaqueline Hess, Jason C Slot and Anne Pringle, dio:10.1093/gbe/evaa193, Genome Biology and Evolution, Nov 2020.
 Convergent Evolution has updates since 2001. Convergent Evolution has updates since 2001.
 ...De Novo Genes has updates since 2009. ...De Novo Genes has updates since 2009.
 Testing Darwinism... suggests that closed-system experiments are better than historical reconstructions. Testing Darwinism... suggests that closed-system experiments are better than historical reconstructions.
 SSoCIA, The Society for Social and Conceptual Issues in Astrobiology holds its 2020 conference beginning next Monday, 14 December. The society deals with questions like, How universal are our moral obligations? Should we attempt to communicate with life beyond our planet? and What is life? Kelly Smith, Chair of the Department of Philosophy & Religion, Clemson University, founded the society in 2016. With international membership, it is small but growing. This year's conference will be entirely virtual – online. We look forward to being there!
SSoCIA, The Society for Social and Conceptual Issues in Astrobiology holds its 2020 conference beginning next Monday, 14 December. The society deals with questions like, How universal are our moral obligations? Should we attempt to communicate with life beyond our planet? and What is life? Kelly Smith, Chair of the Department of Philosophy & Religion, Clemson University, founded the society in 2016. With international membership, it is small but growing. This year's conference will be entirely virtual – online. We look forward to being there!
 About SSoCIA and About SSoCIA and
 President's Welcome. President's Welcome.
| 07 Dec 2020 |
What'sNEW about HGT  | | |
 Mosquito genomes are frequently invaded by transposable elements through horizontal transfer by Elverson Soares de Melo and Gabriel Luz Wallau, doi:10.1371/journal.pgen.1008946, PLoS Genetics, 30 Nov 2020. Mosquito genomes are frequently invaded by transposable elements through horizontal transfer by Elverson Soares de Melo and Gabriel Luz Wallau, doi:10.1371/journal.pgen.1008946, PLoS Genetics, 30 Nov 2020.
 Viruses and Other Gene Transfer Mechanisms has a primer and updates since 1997. Viruses and Other Gene Transfer Mechanisms has a primer and updates since 1997.
...an extreme and inhospitable environment that nonetheless supports microbial life. Even very hot sediments 1,000 meters below the seafloor in an ocean trench off Japan contain viable cells.
 Temperature limits to deep subseafloor life in the Nankai Trough subduction zone by Verena B. Heuer, Fumio Inagaki, Yuki Morono et al., Science, 04 Dec 2020. Temperature limits to deep subseafloor life in the Nankai Trough subduction zone by Verena B. Heuer, Fumio Inagaki, Yuki Morono et al., Science, 04 Dec 2020.
 Bacteria... makes the case that life from space must be able to tolerate extremes. Bacteria... makes the case that life from space must be able to tolerate extremes.
| 27 Nov 2020 |
What'sNEW about HGT  | | |
Green algae contain very many genes from large DNA viruses, we commented last week. Now we learn that DNA polymerase in the same type of virus may share significant homology with eukaryotic DNA polymerase. Under neo-darwinian philosophy, it's hard to say what this might mean. "...He hypothesized that the eukaryotic enzyme originated as a contribution from some ancient poxvirus." (Huh? It existed among viruses before it originated?)
We believe that macroevolutionary progress comes after existing genes are aquired by transfer. If so, eukaryotic DNA polymerase genes in viruses make good sense.
 Did Viruses Create the Nucleus? The Answer May Be Near by Christie Wilcox, Quanta, 25 Nov 2020. Did Viruses Create the Nucleus? The Answer May Be Near by Christie Wilcox, Quanta, 25 Nov 2020.
 26, 24 Sep 2019: RNA polymerases, and tRNAs in large viruses. 26, 24 Sep 2019: RNA polymerases, and tRNAs in large viruses.
 Viruses and Other Gene Transfer Mechanisms has background and updates since 1997. Viruses and Other Gene Transfer Mechanisms has background and updates since 1997.
| 19 Nov 2020 |
What'sNEW about HGT  | | |
In cosmic ancestry, new genetic programs must be supplied by some means of horizontal gene transfer (HGT). Viruses are the most ubiquitous and efficient potential source of supply. Now a new report indicates that nucleocytoplasmic large DNA viruses (NCLDVs) can contibute genes to eukaryotes in quantities previously unimagined.
Here we report the widespread endogenization of NCLDVs in diverse green algae; these giant EVEs [endogenous viral elements] reached sizes greater than 1 million base pairs and contained as many as around 10% of the total open reading frames in some genomes, substantially increasing the scale of known viral genes in eukaryotic genomes. ...Their endogenization represents an underappreciated conduit of new genetic material into eukaryotic lineages that can substantially impact genome composition.
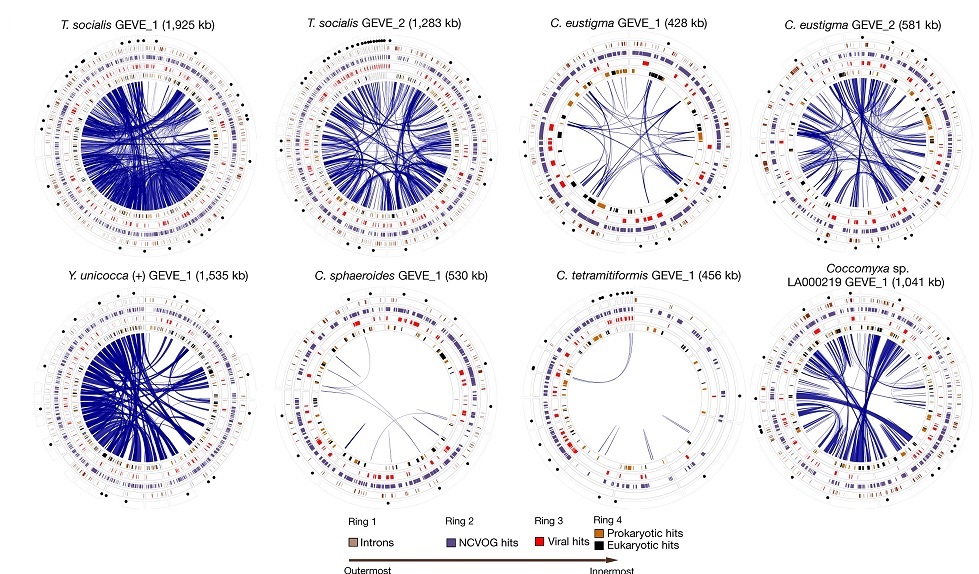
 Widespread endogenization of giant viruses shapes genomes of green algae by M. Moniruzzaman, A.R. Weinheimer, C.A. Martinez-Gutierrez et al., doi:10.1038/s41586-020-2924-2, Nature, 18 Nov 2020. Widespread endogenization of giant viruses shapes genomes of green algae by M. Moniruzzaman, A.R. Weinheimer, C.A. Martinez-Gutierrez et al., doi:10.1038/s41586-020-2924-2, Nature, 18 Nov 2020.
 Giant Viruses Can Integrate into the Genomes of Their Hosts by Amanda Heidt, TheScientist, 19 Nov 2020. Giant Viruses Can Integrate into the Genomes of Their Hosts by Amanda Heidt, TheScientist, 19 Nov 2020.
 Viruses and Other Gene Transfer Mechanisms has background and updates since 1997. Viruses and Other Gene Transfer Mechanisms has background and updates since 1997.
 NASA's Webb Telescope Will Investigate the Intertwined Origins of Dust and Life, Space Telescope Science Institute (+Newswise), 18 Nov 2020. NASA's Webb Telescope Will Investigate the Intertwined Origins of Dust and Life, Space Telescope Science Institute (+Newswise), 18 Nov 2020.
 Hoyle and Wickramasinghe's Analysis of Interstellar Dust has more. Hoyle and Wickramasinghe's Analysis of Interstellar Dust has more.
 Controversy ...over whether phosphine really was discovered on Venus, Physics World, 06 Nov 2020. Controversy ...over whether phosphine really was discovered on Venus, Physics World, 06 Nov 2020.
 Prospects for life on Venus fade–but aren't dead yet by Alexandra Witze, Nature, 17 Nov 2020. Prospects for life on Venus fade–but aren't dead yet by Alexandra Witze, Nature, 17 Nov 2020.
 Thanks, Martin Langford. Thanks, Martin Langford.
 14 Sep 2020: Our notice and links about the phosphene. 14 Sep 2020: Our notice and links about the phosphene.
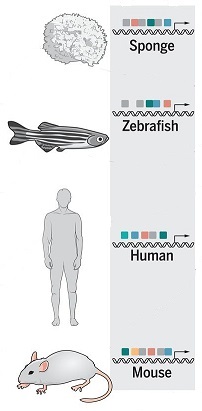 In cosmic ancestry, genomes would have operating systems that can recognize and deploy available programming, even for features previously unexpressed on Earth. New research shows how transfer factors and enhancers shared across the animal kingdom would be components of such systems.
In cosmic ancestry, genomes would have operating systems that can recognize and deploy available programming, even for features previously unexpressed on Earth. New research shows how transfer factors and enhancers shared across the animal kingdom would be components of such systems.
Our results suggest the existence of an ancient and conserved, yet flexible, genomic regulatory syntax that has been repeatedly co-opted into cell type-specific gene regulatory networks across the animal kingdom. ...We posit that the expansion of TFs and enhancers may underlie the evolution of complex body plans.
 Deep conservation of the enhancer regulatory code in animals by Emily S. Wong et al., doi:10.1126/science.aax8137, Science, 06 Nov 2020. Deep conservation of the enhancer regulatory code in animals by Emily S. Wong et al., doi:10.1126/science.aax8137, Science, 06 Nov 2020.
 Robust Software Management in Genomes (under construction) has related comments and links. Robust Software Management in Genomes (under construction) has related comments and links.
Our Galaxy may be teeming with rogue planets, gravitationally unbound to any star.
They are hard to see, but Polish astronomers report spotting one. Could they carry dormant life between stars?
 A Terrestrial-mass Rogue Planet Candidate Detected in the Shortest-timescale Microlensing Event by P. Mróz, R. Poleski, A. Gould et al., Astrophysical Journal Letters, in press | arXiv, last rev., 20 Oct 2020. A Terrestrial-mass Rogue Planet Candidate Detected in the Shortest-timescale Microlensing Event by P. Mróz, R. Poleski, A. Gould et al., Astrophysical Journal Letters, in press | arXiv, last rev., 20 Oct 2020.
 An Earth-sized rogue planet discovered in the Milky Way, University of Warsaw (+Newswise), 29 Oct 2020. An Earth-sized rogue planet discovered in the Milky Way, University of Warsaw (+Newswise), 29 Oct 2020.
Ambitious new research about evolution caught my attention. A team from Spain and the Netherlands used comparative genomics to probe the evolution of eukaryotic cells:
Eukaryogenesis is one of the most enigmatic evolutionary transitions, during which simple prokaryotic cells gave rise to complex eukaryotic cells. While evolutionary intermediates are lacking, gene duplications provide information on the order of events by which eukaryotes originated. Here we use a phylogenomics approach to reconstruct successive steps during eukaryogenesis. ... Altogether, we infer that the host that engulfed the proto-mitochondrion had some eukaryote-like complexity, which drastically increased upon mitochondrial acquisition. This scenario bridges the signs of complexity observed in Asgard archaeal genomes to the proposed role of mitochondria in triggering eukaryogenesis.
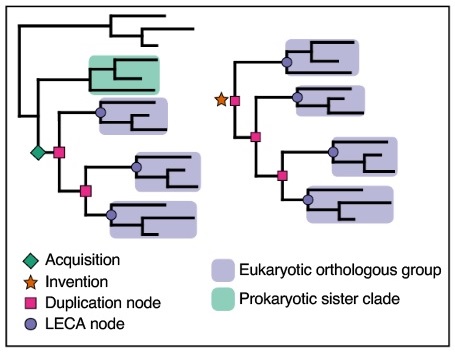 Besides the acquisition of genes via the endosymbiont, the proto-eukaryotic genome expanded through gene inventions, duplications and horizontal gene transfers during eukaryogenesis.
Besides the acquisition of genes via the endosymbiont, the proto-eukaryotic genome expanded through gene inventions, duplications and horizontal gene transfers during eukaryogenesis.
Invention is the phenomenon that I am most curious about. After studying the paper, I wrote to corresponding co-author Dr. Toni Gabaldón of the Comparative Genomics Group at Barcelona Supercomputing Centre, and the Institute for Research in Biomedicine, Barcelona. I asked, "...May I assume that no _process_ of invention was observed, only that the genes had no identifiable predecessors? They seem to 'have come from nowhere?'"
He replied, "Yes is as you say, we call 'invention' those gene families for which predecessors (homologs outside eukaryotes) cannot be identified. Part of these could be families that have diverged beyond recognition, and part of these could be newly originated families. How these families originate?, we do not discuss this. My preferred hypothesis is that they originate de novo from non-coding pieces of the genome as it has been shown that this process can occur in some extant genomes...."
 Timing the origin of eukaryotic cellular complexity with ancient duplications by Julian Vosseberg, Jolien J. E. van Hooff et al., Nature Ecology & Evolution, 26 Oct 2020. Timing the origin of eukaryotic cellular complexity with ancient duplications by Julian Vosseberg, Jolien J. E. van Hooff et al., Nature Ecology & Evolution, 26 Oct 2020.
 Thanks for sharing the full article, Dr. Gabaldón. Thanks for sharing the full article, Dr. Gabaldón.
 Three New Human Genes... has become our main page about de novo genes — genes that already existed, somewhere, as silent sequences, before activation. Three New Human Genes... has become our main page about de novo genes — genes that already existed, somewhere, as silent sequences, before activation.
 19 Feb 2017: recent news about eukaryotic genes in Asgard archaea. 19 Feb 2017: recent news about eukaryotic genes in Asgard archaea.
|



 Three mechanisms of gene family emergence. (A) de novo gene birth; (B) HGT; (C) gene duplication. Blue, species tree; Red, new gene families (arrow indicates origin); Black, homologous genes.
Three mechanisms of gene family emergence. (A) de novo gene birth; (B) HGT; (C) gene duplication. Blue, species tree; Red, new gene families (arrow indicates origin); Black, homologous genes.

 Besides the acquisition of genes via the endosymbiont, the proto-eukaryotic genome expanded through gene inventions, duplications and horizontal gene transfers during eukaryogenesis.
Besides the acquisition of genes via the endosymbiont, the proto-eukaryotic genome expanded through gene inventions, duplications and horizontal gene transfers during eukaryogenesis.