* * * THIS PAGE IS UNDER CONSTRUCTION * * *
Robust Software Management
 What'sNEW What'sNEW
If evolution works by cosmic ancestry, it must depend on robust software management (RSM) within genomes. For an example of this capability, many bacteria are able to respond to environmental threats by forming dormant endospores. The endospore has a multi-layered protective outer coat, has reduced water content, and can resist hazards like starvation, freezing, drought, vacuum, high pressure, acceleration and most poisons. With no metabolism, an endospore may remain viable for millenia, millions of years, possibly indefinitely. And when safe conditions are restored, the cell may return to active life again within minutes. Many genetically directed changes in the cell underlie the conversion from vegetative growth to sporulation (1).
Some of this capability has analogies in our familiar digital world, as when a portable computer runs low on battery power and automatically launches a shutdown routine. But the endospore is more analogous to the computer repackaged into its original protective shipping carton, somehow effected by robust software management.
Some bacteria, like Deinococcus radiodurans, are able to repair their genomes after the DNA has been severly fragmented by radiation. This reminds us of the "Defrag" function on most computers, but with features analogous to syntax- and spell-check included. (1.5)
These features of bacteria are matched by simlar capabilities among eukaryotes, where the genome exists in two nearly identical versions, so there's a backup copy. When a eukaryotic cell repairs a broken gene using the backup version, it's called "gene conversion" (2), an excellent, remarkable example of robust software management. Protective heat shock proteins (in prokaryotes as well) are quickly produced when the cell experiences a sudden rise in temperature. (2.5) More examples of impressive software management include double strand break repair, gene duplication, the generation of intronless paralogs, and, certainly, meiosis.
Changing Environments
The examples given so far mainly pertain to protecting the cell or species and keeping the genome uncorrupted. But sometimes, in new situations, genetic changes are needed. There is plenty of evidence that genetic programs can be optimized to suit changed conditions. A familiar example is the color vision of coelacanths, living 200 meters underwater where only dim blue light is available. In each of two color-receptors, only two amino acids are changed from the orthologous receptors in species living in brighter light. Each of these changes could be accomplished by one nucleotide substitution, not forbiddingly unlikely. Examples of similar optimization are everywhere in the tree of life.
Of course, random nucleotide substitutions are usually harmful and sometimes fatal. Therefore, it would be better if the tinkering were supressed until a need arises. There is programming to initiate the tinkering. The phenomenon is called "adaptive mutation." By one account, ...the newly identified mutases, present in all cells, produce mutations only when a genetic or metabolic stress triggers their induction and activation. (2.6)
It would also help if the mutations were focussed on the appropriate nucleotides only, the ones needing to change. Indeed, "directed mutation" often confines the point mutations to positions where they may be useful. Among prokaryotes,
diversity-generating retroelements (DGRs) use mutagenic reverse transcription and retrohoming to generate myriad variants of a target gene. ...Crucially, the reverse transcriptase (RT) used is error-prone at template adenine bases, but has high fidelity at other template bases.... Massive and low-risk protein diversification offers clear advantages to any organism. (4.5)
Macroevolution
Adaptation to changing environments often requires only microevolution — evolution attainable with minor tweaking and optimizing of existing proteins. Macroevolution, by contrast, requires wholly new programs or subroutines. It is best illustrated by example. Examples would include the first earthly appearances of [a long list]. These evolutionary features depend on the first deployment of genetic sequences that contain the programming for [the long list]. This first deployment requires (1) that the programming is available, and (2) that the regulatory system is appropriate for it and synchronized with it. But where does the programming come from?
Horizontal Gene Transfer
is the whole story among bacteria
can be accelerated
can be initiated by the recipient species (5). bacteria can kill to steal
"the amoeba replaced it with another gene with the same function from bacteria." (4)
viral infection can transform whole eukaryotic species in few generations (3.5)
| The complex capabilities exemplified here are made possible by lengthy genetic programming. Today's theory of evolution attempts to explain how the programs originated. But the explanations are missing and the origins are only assumed. |
Adaptive / Directed Mutation
Regulatory Changes
Enhancer Grammar? (6)
Reverse Transcription
Intron "Homing"
Lagging Strand Replication via Okazaki Fragments
Pseudogenes
Gene Conversion
Meiosis
"...if there is one event in the whole evolutionary sequence at which my own mind lets my awe still overcome my instinct to analyse, and where I might concede that there may be a difficulty in seeing a Darwinian gradualism hold sway throughout almost all, it is this event–the initiation of meiosis." W. J. Hamilton, Narrow Roads to Gene Land: Evolution of Sex, Vol. 2., Oxford University Press, 1999, quoted in "The Evolution of Meiosis From Mitosis," by Adam S Wilkins and Robin Holliday, Genetics, 01 Jan 2009.
Metamorphosis
Diversity Generating Retroelements
What'sNEW
—
including genetic surprises that look like aspects of RSM
 11 Apr 2025: ...complete, phased, diploid genomes of six ape species.... 11 Apr 2025: ...complete, phased, diploid genomes of six ape species....
 "Telomeric transposons are pervasive in linear bacterial genomes," by Shan-Chi Hsieh et al, Science, 28 Mar 2025.
One subfamily of telomeric transposons co-opted a CRISPR system ...as a tool for the transposon to target chromosome ends, exploiting RNA guides to locate and target telomeric sequences. "Telomeric transposons are pervasive in linear bacterial genomes," by Shan-Chi Hsieh et al, Science, 28 Mar 2025.
One subfamily of telomeric transposons co-opted a CRISPR system ...as a tool for the transposon to target chromosome ends, exploiting RNA guides to locate and target telomeric sequences.
 "How do tardigrades survive intense radiation?" by Bob Goldstein, Current Biology, 24 Mar 2025.
...a protein that can concentrate DNA repair machinery at sites of DNA damage. "How do tardigrades survive intense radiation?" by Bob Goldstein, Current Biology, 24 Mar 2025.
...a protein that can concentrate DNA repair machinery at sites of DNA damage.
 "RNA control of reverse transcription in a diversity-generating retroelement," by Handa, S., Biswas, T., Chakraborty, J. et al, doi:10.1038/s41586-024-08405-w, Nature, online 08 Jan 2025.
The words evolve and evolution are nowhere in the text. Instead the authors notice extraordinary conservation. "RNA control of reverse transcription in a diversity-generating retroelement," by Handa, S., Biswas, T., Chakraborty, J. et al, doi:10.1038/s41586-024-08405-w, Nature, online 08 Jan 2025.
The words evolve and evolution are nowhere in the text. Instead the authors notice extraordinary conservation.
 "Transposon-host arms race: a saga of genome evolution," by YW Iwasaki et al, doi:10.1016/j.tig.2025.01.009, TiG, online 20 Feb 2025.
...arms race? or testing and selection by robust software management (RSM)? "Transposon-host arms race: a saga of genome evolution," by YW Iwasaki et al, doi:10.1016/j.tig.2025.01.009, TiG, online 20 Feb 2025.
...arms race? or testing and selection by robust software management (RSM)?
 "Species-specific circular RNA circDS-1 enhances adaptive evolution...," by Xueyan Hu et al, doi:10.1371/journal.pgen.1011482, PLoS Genet., 08 Mar 2025.
...a novel molecular mechanism for the adaptive evolution of functional circRNAs derived from intronic mutations. "Species-specific circular RNA circDS-1 enhances adaptive evolution...," by Xueyan Hu et al, doi:10.1371/journal.pgen.1011482, PLoS Genet., 08 Mar 2025.
...a novel molecular mechanism for the adaptive evolution of functional circRNAs derived from intronic mutations.
 "Nucleosome fibre topology guides transcription factor binding to enhancers," by Michael R. O'Dwyer et al, Nature, 06 Feb 2025. "Nucleosome fibre topology guides transcription factor binding to enhancers," by Michael R. O'Dwyer et al, Nature, 06 Feb 2025.
 "Chromatin-centric insights into DNA replication," by Yang Liu et al, doi:10.1016/j.tig.2024.12.003, TiG, 07 Jan 2025. "Chromatin-centric insights into DNA replication," by Yang Liu et al, doi:10.1016/j.tig.2024.12.003, TiG, 07 Jan 2025.
 "Conserved features of recombination control in vertebrates," by Linda Odenthal-Hesse, doi:10.1371/journal.pbio.3002959, PLoS Biol., 07 Jan 2025. "Conserved features of recombination control in vertebrates," by Linda Odenthal-Hesse, doi:10.1371/journal.pbio.3002959, PLoS Biol., 07 Jan 2025.
 "...No-go decay plays a role during embryo development in zebrafish," by Alana R. Plastrik and Hani S. Zaher, doi:10.1371/journal.pbio.3002925, PLoS Biol., 06 Dec 2024. "...No-go decay plays a role during embryo development in zebrafish," by Alana R. Plastrik and Hani S. Zaher, doi:10.1371/journal.pbio.3002925, PLoS Biol., 06 Dec 2024.
 "Transposable element exonization generates a reservoir of evolving and functional protein isoforms," by Marianne Burbage, Antonela Merlotti, Sebastian Amigorena et al, doi:10.1016/j.cell.2024.11.011, Cell, online 11 Dec 2024. "Transposable element exonization generates a reservoir of evolving and functional protein isoforms," by Marianne Burbage, Antonela Merlotti, Sebastian Amigorena et al, doi:10.1016/j.cell.2024.11.011, Cell, online 11 Dec 2024.
 18 Nov 2024: Transferred genes likely brought along their own regulatory elements. 18 Nov 2024: Transferred genes likely brought along their own regulatory elements.
 "The ribosome comes to life," by Harry F. Noller, Cell, 14 Nov 2024. "The ribosome comes to life," by Harry F. Noller, Cell, 14 Nov 2024.
 "Two-factor authentication underpins the precision of the piRNA pathway," by Dias Mirandela, M., Zoch, A., Leismann, J. et al, Nature, 18 Sep 2024.
We propose that piRNA-directed LINE1 DNA methylation requires a developmentally timed two-factor authentication process. "Two-factor authentication underpins the precision of the piRNA pathway," by Dias Mirandela, M., Zoch, A., Leismann, J. et al, Nature, 18 Sep 2024.
We propose that piRNA-directed LINE1 DNA methylation requires a developmentally timed two-factor authentication process.
 "Intragenc DNA inversions expand bacterial coding capacity," by Rachael B. Chanin, Patrick T. West et al, Nature, 25 Sep 2024.
Bacteria can generate heterogeneity through phase variation – a preorogrammed, reversible mechanism that alters gene level expression across a population. ...We suspect that there is an underlying 'molecular grammar'.... ...invertases may be expressed in response to specific cues or signals. ...Additionally, we anticipate that future studies of intragenic inversion will uncover new layers of bioregulation in prokaryotes and demonstrate many hidden programmes that exist within highly plastic bacterial genomes. "Intragenc DNA inversions expand bacterial coding capacity," by Rachael B. Chanin, Patrick T. West et al, Nature, 25 Sep 2024.
Bacteria can generate heterogeneity through phase variation – a preorogrammed, reversible mechanism that alters gene level expression across a population. ...We suspect that there is an underlying 'molecular grammar'.... ...invertases may be expressed in response to specific cues or signals. ...Additionally, we anticipate that future studies of intragenic inversion will uncover new layers of bioregulation in prokaryotes and demonstrate many hidden programmes that exist within highly plastic bacterial genomes.
 "Sensing DNA as danger: The discovery of cGAS," by Richard A. Flavell1 and Esen Sefik, Immunity, 08 Oct 2024.
cGAS binds to DNA, which confers on it the ability to synthesize cyclic GMP-AMP (cGAMP) that in turn enables stimulator of interferon (IFN) genes (STING) to activate the IFN response through the transcription factor IRF3. "Sensing DNA as danger: The discovery of cGAS," by Richard A. Flavell1 and Esen Sefik, Immunity, 08 Oct 2024.
cGAS binds to DNA, which confers on it the ability to synthesize cyclic GMP-AMP (cGAMP) that in turn enables stimulator of interferon (IFN) genes (STING) to activate the IFN response through the transcription factor IRF3.
 "Exploring the world of small proteins in plant biology and bioengineering," by Louise Petri et al, TiG, 14 Oct 2024. "Exploring the world of small proteins in plant biology and bioengineering," by Louise Petri et al, TiG, 14 Oct 2024.
 10 Oct 2024: Micro-RNAs are seen to play a role in gene regulation. 10 Oct 2024: Micro-RNAs are seen to play a role in gene regulation.
 "Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production," by Anabel Rodriguez et al, PNAS, 27 Aug 2024. "Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production," by Anabel Rodriguez et al, PNAS, 27 Aug 2024.
 "Structural basis of human 20S proteasome biogenesis," by Zhang, H., Zhou, C., Mohammad, Z. et al, Nat Commun; and commentary: PhysOrg, 21 Sep 2024. "Structural basis of human 20S proteasome biogenesis," by Zhang, H., Zhou, C., Mohammad, Z. et al, Nat Commun; and commentary: PhysOrg, 21 Sep 2024.
 "Programmed DNA elimination," by Kazufumi Mochizuki, Current Biology, 23 Sep 2024. "Programmed DNA elimination," by Kazufumi Mochizuki, Current Biology, 23 Sep 2024.
 "FANCD2–FANCI surveys DNA and recognizes double- to single-stranded junctions," by Pablo Alcón et al, Nature, 29 Aug 2024. "FANCD2–FANCI surveys DNA and recognizes double- to single-stranded junctions," by Pablo Alcón et al, Nature, 29 Aug 2024.
 "Distinct roles of spindle checkpoint proteins in meiosis," by Anuradha Mukherjee et al, Current Biology, 29 Jul 2024. "Distinct roles of spindle checkpoint proteins in meiosis," by Anuradha Mukherjee et al, Current Biology, 29 Jul 2024.
 "The molecular architecture of the nuclear basket," by Digvijay Singh,
Neelesh Soni, Joshua Hutchings et al, Cell, 09 Aug 2024. ("The light of evolution" illuminates nothing here; the word does not appear in the article.) "The molecular architecture of the nuclear basket," by Digvijay Singh,
Neelesh Soni, Joshua Hutchings et al, Cell, 09 Aug 2024. ("The light of evolution" illuminates nothing here; the word does not appear in the article.)
 "Single-molecule imaging reveals the mechanism of bidirectional replication initiation in metazoa," by Riki Terui et al, doi:10.1016/j.cell.2024.05.024, Cell, 11 Jun 2024.
...replication initiation is surprisingly dynamic. "Single-molecule imaging reveals the mechanism of bidirectional replication initiation in metazoa," by Riki Terui et al, doi:10.1016/j.cell.2024.05.024, Cell, 11 Jun 2024.
...replication initiation is surprisingly dynamic.
 "Bdelloid rotifers deploy horizontally acquired biosynthetic genes against a fungal pathogen," by Nowell, R.W., Rodriguez, F., Hecox-Lea, B.J. et al, doi:10.1038/s41467-024-49919-1, Nat Commun; and commentary from Marine Biological Laboratory via EurekAlert!, 18 Jul 2024.
...horizontally acquired genes were over twice as likely to be upregulated as other genes.... "Bdelloid rotifers deploy horizontally acquired biosynthetic genes against a fungal pathogen," by Nowell, R.W., Rodriguez, F., Hecox-Lea, B.J. et al, doi:10.1038/s41467-024-49919-1, Nat Commun; and commentary from Marine Biological Laboratory via EurekAlert!, 18 Jul 2024.
...horizontally acquired genes were over twice as likely to be upregulated as other genes....
 "Non-canonical functions of enhancers: regulation of RNA polymerase III transcription, DNA replication, and V(D)J recombination," by Kevin Struhl, doi:10.1016/j.tig.2024.04.001, TiG, 19 Apr 2024. "Non-canonical functions of enhancers: regulation of RNA polymerase III transcription, DNA replication, and V(D)J recombination," by Kevin Struhl, doi:10.1016/j.tig.2024.04.001, TiG, 19 Apr 2024.
 30 May 2024: GTAs are intricately regulated domesticated viruses that package host DNA into virus-like capsids and transfer this DNA.... 30 May 2024: GTAs are intricately regulated domesticated viruses that package host DNA into virus-like capsids and transfer this DNA....
 "Dual-role transcription factors stabilize intermediate expression levels," by Jinnan He, Xiangru Huo, Gaofeng Pei, Zeran Jia et al., doi:10.1016/j.cell.2024.03.023, Cell, 16 Apr 2024. "Dual-role transcription factors stabilize intermediate expression levels," by Jinnan He, Xiangru Huo, Gaofeng Pei, Zeran Jia et al., doi:10.1016/j.cell.2024.03.023, Cell, 16 Apr 2024.
 "Structural basis of DNA crossover capture by Escherichia coli DNA gyrase," by Marlène Vayssières et al., doi:10.1126/science.adl5899, Science, 11 Apr 2024. "Structural basis of DNA crossover capture by Escherichia coli DNA gyrase," by Marlène Vayssières et al., doi:10.1126/science.adl5899, Science, 11 Apr 2024.
 "Gene regulation during meiosis," by Jingyi Gao, Yiwen Qin and John C. Schimenti, TiG, 03 Jan 2024. "Gene regulation during meiosis," by Jingyi Gao, Yiwen Qin and John C. Schimenti, TiG, 03 Jan 2024.
 "Sister chromatid cohesion establishment during DNA replication termination," by George Cameron et al., Science, 14 Mar 2024. "Sister chromatid cohesion establishment during DNA replication termination," by George Cameron et al., Science, 14 Mar 2024.
 "Scientists discover a key quality-control mechanism in DNA replication," the Perelman School of Medicine at the University of Pennsylvania via PhysOrg, 29 Mar 2024.
...a multi-protein "machine" in cells that helps govern the pausing or stopping of DNA replication to ensure its smooth progress. "Scientists discover a key quality-control mechanism in DNA replication," the Perelman School of Medicine at the University of Pennsylvania via PhysOrg, 29 Mar 2024.
...a multi-protein "machine" in cells that helps govern the pausing or stopping of DNA replication to ensure its smooth progress.

 "Parental histone transfer caught at the replication fork," by Ningning Li, Yuan Gao, Yujie Zhang, Daqi Yu et al., doi:10.1038/s41586-024-07152-2, Nature, 28 Mar 2024.
Our findings offer crucial structural insights into the mechanism of replication-coupled histone recycling for maintaining epigenetic inheritance. "Parental histone transfer caught at the replication fork," by Ningning Li, Yuan Gao, Yujie Zhang, Daqi Yu et al., doi:10.1038/s41586-024-07152-2, Nature, 28 Mar 2024.
Our findings offer crucial structural insights into the mechanism of replication-coupled histone recycling for maintaining epigenetic inheritance.
 "FIRRM and FIGNL1: partners in the regulation of homologous recombination," by Stavroula Tsaridou, Marcel A.T.M. van Vugt et al., doi:10.1016/j.tig.2024.02.007, TiG, 16 Mar 2024.
DNA repair through homologous recombination (HR) is of vital importance for maintaining genome stability and preventing tumorigenesis. RAD51 is the core component of HR, catalyzing the strand invasion and homology search. "FIRRM and FIGNL1: partners in the regulation of homologous recombination," by Stavroula Tsaridou, Marcel A.T.M. van Vugt et al., doi:10.1016/j.tig.2024.02.007, TiG, 16 Mar 2024.
DNA repair through homologous recombination (HR) is of vital importance for maintaining genome stability and preventing tumorigenesis. RAD51 is the core component of HR, catalyzing the strand invasion and homology search.
 "Translation selectively destroys non-functional transcription complexes," by J. Woodgate, H. Mosaei, P. Brazda et al., doi:10.1038/s41586-023-07014-3, Nature, 22 Feb 2024. "Translation selectively destroys non-functional transcription complexes," by J. Woodgate, H. Mosaei, P. Brazda et al., doi:10.1038/s41586-023-07014-3, Nature, 22 Feb 2024.
 "A new family of bacterial ribosome hibernation factors," by Karla Helena-Bueno, Mariia Yu. Rybak at al, https://doi.org/10.1038/s41586-024-07041-8, Nature, 29 Feb 2024. "A new family of bacterial ribosome hibernation factors," by Karla Helena-Bueno, Mariia Yu. Rybak at al, https://doi.org/10.1038/s41586-024-07041-8, Nature, 29 Feb 2024.
 "Persistence of backtracking by human RNA polymerase II," by Kevin B. Yang et al., doi:10.1016/j.molcel.2024.01.019, Molecular Cell, 09 Feb 2024. "Persistence of backtracking by human RNA polymerase II," by Kevin B. Yang et al., doi:10.1016/j.molcel.2024.01.019, Molecular Cell, 09 Feb 2024.
 20 Jan 2024: Evidence for deterministic evolution.... 20 Jan 2024: Evidence for deterministic evolution....
 04 Jan 2024: How periwinkles became viviparous — apparently, the programming was available and the perinkle genome was able to piece it together. 04 Jan 2024: How periwinkles became viviparous — apparently, the programming was available and the perinkle genome was able to piece it together.
 "Power-law behavior of transcriptional bursting regulated by enhancer-promoter communication," by Zihao Wang et al., Genome Res., 03 Jan 2024. "Power-law behavior of transcriptional bursting regulated by enhancer-promoter communication," by Zihao Wang et al., Genome Res., 03 Jan 2024.
 "Structural visualization of transcription initiation in action," by Xizi Chen et al., Science, 22 Dec 2023.
...structural visualization of transcription initiation in action.... "Structural visualization of transcription initiation in action," by Xizi Chen et al., Science, 22 Dec 2023.
...structural visualization of transcription initiation in action....
 "Molecular determinants of ligand efficacy and potency in GPCR signaling," by Franziska M. Heydenreich et al., Science, 22 Dec 2023.
CA Comments "Molecular determinants of ligand efficacy and potency in GPCR signaling," by Franziska M. Heydenreich et al., Science, 22 Dec 2023.
CA Comments
 "Mice possess natural gene therapy system," Hokkaido University (+Newswise), 14 Dec 2023.
...a naturally produced RNA that can regulate alternative splicing in a definitive on/off manner. ...a substantial portion of such non-coding RNAs may be involved in controlling alternative splicing. "Mice possess natural gene therapy system," Hokkaido University (+Newswise), 14 Dec 2023.
...a naturally produced RNA that can regulate alternative splicing in a definitive on/off manner. ...a substantial portion of such non-coding RNAs may be involved in controlling alternative splicing.
 "Generation of de novo miRNAs from template switching during DNA replication," by Heli A. M. Mönttinen et al., PNAS, 29 Nov 2023.
...template switching ...allows for near-instant rewiring of genetic information.... "Generation of de novo miRNAs from template switching during DNA replication," by Heli A. M. Mönttinen et al., PNAS, 29 Nov 2023.
...template switching ...allows for near-instant rewiring of genetic information....
 "rDNA magnification is a unique feature of germline stem cells," by Jonathan O. Nelson et al., PNAS, 15 Nov 2023.
rDNA and telomere likely constitute two major genomic elements that require active maintenance in the germline. "rDNA magnification is a unique feature of germline stem cells," by Jonathan O. Nelson et al., PNAS, 15 Nov 2023.
rDNA and telomere likely constitute two major genomic elements that require active maintenance in the germline.
 "tRNA-derived small RNAs are embedded in the gene regulatory program instructing Drosophila metamorphosis," by Junling Shi, Jiaqi Xu, Jun Ma and Feng He, Genome Res., 16 Nov 2023.
We show that specific tsRNAs indeed exhibit a dynamic accumulation upon entering the pupal stage. "tRNA-derived small RNAs are embedded in the gene regulatory program instructing Drosophila metamorphosis," by Junling Shi, Jiaqi Xu, Jun Ma and Feng He, Genome Res., 16 Nov 2023.
We show that specific tsRNAs indeed exhibit a dynamic accumulation upon entering the pupal stage.
 "Five families of diverse DNA viruses comprehensively restructure the nucleus," by Quincy Rosemarie and Bill Sugden, PLoS Biol., 06 Nov 2023.
Given the ubiquity of virus-induced ROCC, a greater understanding will likely lead to further insights into how the viruses that induce it manipulate their hosts, and how to intervene therapeutically in their life cycles. "Five families of diverse DNA viruses comprehensively restructure the nucleus," by Quincy Rosemarie and Bill Sugden, PLoS Biol., 06 Nov 2023.
Given the ubiquity of virus-induced ROCC, a greater understanding will likely lead to further insights into how the viruses that induce it manipulate their hosts, and how to intervene therapeutically in their life cycles.
 10 Nov 2023: ...much of the "dark matter" transcriptome may be a by-product, or even an expected component, of the regulation of known genes, as well as a source of novel genetic entities. 10 Nov 2023: ...much of the "dark matter" transcriptome may be a by-product, or even an expected component, of the regulation of known genes, as well as a source of novel genetic entities.
 "Pioneer factors: roles and their regulation in development," by Amandine Barral and Kenneth S. Zaret, Trends in Genetics, 06 Nov 2023. "Pioneer factors: roles and their regulation in development," by Amandine Barral and Kenneth S. Zaret, Trends in Genetics, 06 Nov 2023.
 "Evolutionary innovation through transcription factor rewiring in microbes...," by Matthew J. Shepherd et al., PLoS Biol, 23 Oct 2023.
...we reveal a hierarchy among transcription factors that are rewired to rescue lost function, with alternative rewiring pathways only unmasked after the preferred pathway is eliminated. "Evolutionary innovation through transcription factor rewiring in microbes...," by Matthew J. Shepherd et al., PLoS Biol, 23 Oct 2023.
...we reveal a hierarchy among transcription factors that are rewired to rescue lost function, with alternative rewiring pathways only unmasked after the preferred pathway is eliminated.
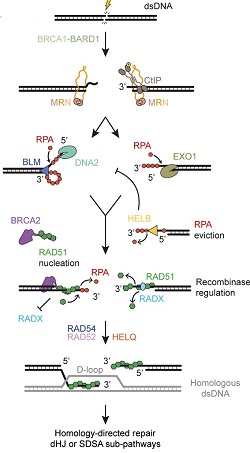
 "Recent insights into eukaryotic double-strand DNA break repair...," by Sara De Bragança et al., TiG, 06 Oct 2023.
Both pathways involve complex DNA transactions catalyzed by proteins that sequentially or cooperatively work to repair the damage. (Homologous recombination illustrated at right, click to enlarge.) "Recent insights into eukaryotic double-strand DNA break repair...," by Sara De Bragança et al., TiG, 06 Oct 2023.
Both pathways involve complex DNA transactions catalyzed by proteins that sequentially or cooperatively work to repair the damage. (Homologous recombination illustrated at right, click to enlarge.)
 "Self-demixing of mRNA copies buffers mRNA:mRNA and mRNA:regulator stoichiometries," by Andrés H. Cardona, Szilvia Ecsedi et al., Cell, 12 Sep 2023.
Robust control of mRNA:mRNA and mRNA:protein stoichiometries emerges from mRNA self-demixing and cooperative super-assembly into multiphase multiscale condensates with dynamic storage capacity. "Self-demixing of mRNA copies buffers mRNA:mRNA and mRNA:regulator stoichiometries," by Andrés H. Cardona, Szilvia Ecsedi et al., Cell, 12 Sep 2023.
Robust control of mRNA:mRNA and mRNA:protein stoichiometries emerges from mRNA self-demixing and cooperative super-assembly into multiphase multiscale condensates with dynamic storage capacity.
 "Ultra-long-range interactions between active regulatory elements," by Elias T. Friman et al., Genome Res., online 14 Jul 2023. "Ultra-long-range interactions between active regulatory elements," by Elias T. Friman et al., Genome Res., online 14 Jul 2023.
 "Transposable elements in mammalian chromatin organization," by H.A. Lawson, Y. Liang and T. Wang, Nat Rev Genet, Oct 2023. "Transposable elements in mammalian chromatin organization," by H.A. Lawson, Y. Liang and T. Wang, Nat Rev Genet, Oct 2023.
 "Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity," by A. Wenger, A. Biran, N. Alcaraz et al., Nat Genet, 04 Sep 2023. "Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity," by A. Wenger, A. Biran, N. Alcaraz et al., Nat Genet, 04 Sep 2023.
 "Evelyn M. Witkin (1921–2023): Pioneer of cell mutagenesis and DNA repair research," by Joann B. Sweasy and Philip C. Hanawalt, Science, 07 Sep 2023. "Evelyn M. Witkin (1921–2023): Pioneer of cell mutagenesis and DNA repair research," by Joann B. Sweasy and Philip C. Hanawalt, Science, 07 Sep 2023.
 "Pervasive downstream RNA hairpins dynamically dictate start-codon selection," by Y. Xiang, W. Huang, L. Tan et al., Nature, 29 Jul 2023. "Pervasive downstream RNA hairpins dynamically dictate start-codon selection," by Y. Xiang, W. Huang, L. Tan et al., Nature, 29 Jul 2023.
 "Membrane-associated DNA transport machines," by Briana Burton and David Dubnau, Cold Spring Harb Perspect Biol., Jul 2010. "Membrane-associated DNA transport machines," by Briana Burton and David Dubnau, Cold Spring Harb Perspect Biol., Jul 2010.
 "Pre-mRNA splicing and its cotranscriptional connections," by Hossein Shenasa and David L. Bentley, Cell, 24 May 2023. "Pre-mRNA splicing and its cotranscriptional connections," by Hossein Shenasa and David L. Bentley, Cell, 24 May 2023.
 "The circadian clock of the bacterium B. subtilis evokes properties of complex, multicellular circadian systems," by Francesca Sartor et al., Science Advances, 04 Aug 2023. "The circadian clock of the bacterium B. subtilis evokes properties of complex, multicellular circadian systems," by Francesca Sartor et al., Science Advances, 04 Aug 2023.
 "Orphan quality control shapes network dynamics and gene expression," by Kevin G. Mark, SriDurgaDevi Kolla, Jacob D. Aguirre et al., Cell, 20 Jul 2023. "Orphan quality control shapes network dynamics and gene expression," by Kevin G. Mark, SriDurgaDevi Kolla, Jacob D. Aguirre et al., Cell, 20 Jul 2023.
 29 Jun 2023: Eukaryotes appear to have a feature analogous to the CRISPR gene editing system in bacteria. 29 Jun 2023: Eukaryotes appear to have a feature analogous to the CRISPR gene editing system in bacteria.
 "The human embryo selection arena is associated with transposable element activity," by Anna Osnato et al., PLoS Biol., 20 Jun 2023. "The human embryo selection arena is associated with transposable element activity," by Anna Osnato et al., PLoS Biol., 20 Jun 2023.
 "Mitotic bookmarking by SWI/SNF subunits," by Z. Zhu, X. Chen, A. Guo et al., Nature, 24 May 2023. "Mitotic bookmarking by SWI/SNF subunits," by Z. Zhu, X. Chen, A. Guo et al., Nature, 24 May 2023.
 15 May 2023: Concerted Evolution? 15 May 2023: Concerted Evolution?
 02 Apr 2023: Introns and Introners. 02 Apr 2023: Introns and Introners.
 "De novo genes with an lncRNA origin encode unique human brain developmental functionality," by N.A. An, J. Zhang, F. Mo et al., doi:10.1038/s41559-022-01925-6, Nat Ecol Evol, 02 Jan 2023. "De novo genes with an lncRNA origin encode unique human brain developmental functionality," by N.A. An, J. Zhang, F. Mo et al., doi:10.1038/s41559-022-01925-6, Nat Ecol Evol, 02 Jan 2023.
 "Remodeling of maternal mRNA through poly(A) tail orchestrates human oocyte-to-embryo transition," by Yusheng Liu, Han Zhao, Fanghong Shao, Yiwei Zhang et al., OA link, Nature Structural & Molecular Biology, 16 Jan 2023 | my comments, 13 Feb 2023. "Remodeling of maternal mRNA through poly(A) tail orchestrates human oocyte-to-embryo transition," by Yusheng Liu, Han Zhao, Fanghong Shao, Yiwei Zhang et al., OA link, Nature Structural & Molecular Biology, 16 Jan 2023 | my comments, 13 Feb 2023.
 ATRX, a guardian of chromatin, by Paula Aguilera and Andrés J. López-Contreras, doi:10.1016/j.tig.2023.02.009, TiG, 07 Mar 2023.
ATRX limits the formation of secondary structures that can trigger replication fork stalling and replication stress. Furthermore, ATRX regulates DNA repair via homologous recombination. ATRX, a guardian of chromatin, by Paula Aguilera and Andrés J. López-Contreras, doi:10.1016/j.tig.2023.02.009, TiG, 07 Mar 2023.
ATRX limits the formation of secondary structures that can trigger replication fork stalling and replication stress. Furthermore, ATRX regulates DNA repair via homologous recombination.
 Noncanonical DNA structures are drivers of genome evolution, by Kateryna D. Makova and Matthias H. Weissensteiner, doi:10.1016/j.tig.2022.11.005, TiG, 03 Jan 2023. Noncanonical DNA structures are drivers of genome evolution, by Kateryna D. Makova and Matthias H. Weissensteiner, doi:10.1016/j.tig.2022.11.005, TiG, 03 Jan 2023.
 Towards a unification of the 2 meanings of 'epigenetics', by Sui Hwang, doi:10.1371/journal.pbio.3001944, PLoS Biol, 27 Dec 2022. Towards a unification of the 2 meanings of 'epigenetics', by Sui Hwang, doi:10.1371/journal.pbio.3001944, PLoS Biol, 27 Dec 2022.
 Human evolution wasn't just the sheet music, but how it was played (commentary on HAQERS research, next below), from Duke University via Newswise, 23 Nov 2022. Human evolution wasn't just the sheet music, but how it was played (commentary on HAQERS research, next below), from Duke University via Newswise, 23 Nov 2022.
 Adaptive sequence divergence forged new neurodevelopmental enhancers in humans, by Riley J. Mangan et al., doi:10.1016/j.cell.2022.10.016, Cell, 23 Nov 2022. HAQERs evolved under elevated mutation rates and positive selection. Adaptive sequence divergence forged new neurodevelopmental enhancers in humans, by Riley J. Mangan et al., doi:10.1016/j.cell.2022.10.016, Cell, 23 Nov 2022. HAQERs evolved under elevated mutation rates and positive selection.
 Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation, by Jinfan Wang et al., doi:10.1016/j.cell.2022.10.005, Cell, 04 Nov 2022. Scanning occurs at ~100 nucleotides per second. Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation, by Jinfan Wang et al., doi:10.1016/j.cell.2022.10.005, Cell, 04 Nov 2022. Scanning occurs at ~100 nucleotides per second.
 09 Oct 2022: Bacterial spore's membrane acts as a "biological capacitor." 09 Oct 2022: Bacterial spore's membrane acts as a "biological capacitor."
 Transperons: RNA operons as effectors of coordinated gene expression in eukaryotes, by Rohini R. Nair et al., doi:10.1016/j.tig.2022.07.005, TiG, 04 Aug 2022. RNA operons ...regulate coordinated gene expression. ...mRNAs derived from different chromosomes assemble into ribonucleoprotein particles (RNPs) that act as functional operons to generate protein cohorts upon cotranslation. Transperons: RNA operons as effectors of coordinated gene expression in eukaryotes, by Rohini R. Nair et al., doi:10.1016/j.tig.2022.07.005, TiG, 04 Aug 2022. RNA operons ...regulate coordinated gene expression. ...mRNAs derived from different chromosomes assemble into ribonucleoprotein particles (RNPs) that act as functional operons to generate protein cohorts upon cotranslation.
 25 years of the segmentation clock gene, by Ryoichiro Kageyama, doi:10.1038/d41586-022-03562-2, Nature, 09 Nov 2022. 25 years of the segmentation clock gene, by Ryoichiro Kageyama, doi:10.1038/d41586-022-03562-2, Nature, 09 Nov 2022.
 Single-particle studies of the effects of RNA protein interactions on the self-assembly of RNA virus particles, by Rees F Garmann et al., doi:10.1073/pnas.2206292119, PNAS, 19 Sep 2022. The nucleated pathway observed with the plant virus BMV is strikingly similar to that previously observed with bacteriophage MS2, a phylogenetically distinct virus with a different host kingdom. These results raise the possibility that nucleated assembly pathways might be common to other RNA viruses. Single-particle studies of the effects of RNA protein interactions on the self-assembly of RNA virus particles, by Rees F Garmann et al., doi:10.1073/pnas.2206292119, PNAS, 19 Sep 2022. The nucleated pathway observed with the plant virus BMV is strikingly similar to that previously observed with bacteriophage MS2, a phylogenetically distinct virus with a different host kingdom. These results raise the possibility that nucleated assembly pathways might be common to other RNA viruses.
 Group II intron-like reverse transcriptases function in double-strand break repair, by Seung Kuk Park et al., doi:10.1016/j.cell.2022.08.014, Cell, online 15 Sep 2022. Group II intron-like reverse transcriptases function in double-strand break repair, by Seung Kuk Park et al., doi:10.1016/j.cell.2022.08.014, Cell, online 15 Sep 2022.
 Nested epistasis enhancer networks for robust genome regulation, by Xueqiu Lin et al., doi:10.1126/science.abk3512, Science, 11 Aug 2022. Nested epistasis enhancer networks for robust genome regulation, by Xueqiu Lin et al., doi:10.1126/science.abk3512, Science, 11 Aug 2022.
 Correspondence with James Shapiro mentions Natural Genetic Engineering (NGE), Jun-Sep 2022. Correspondence with James Shapiro mentions Natural Genetic Engineering (NGE), Jun-Sep 2022.
 "LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs" by Jun-Yan Kang et al., doi:10.1126/science.abj6647, Science, 12 Aug 2022. "LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs" by Jun-Yan Kang et al., doi:10.1126/science.abj6647, Science, 12 Aug 2022.
 "Expression Plasticity of Transposable Elements Is Highly Associated with Organismal Re-adaptation to Ancestral Environments" by Yan-Nan Liu et al., doi:10.1093/gbe/evac084, Genome Biology and Evolution, 01 Jun 2022. "Expression Plasticity of Transposable Elements Is Highly Associated with Organismal Re-adaptation to Ancestral Environments" by Yan-Nan Liu et al., doi:10.1093/gbe/evac084, Genome Biology and Evolution, 01 Jun 2022.
 "Genome surveillance by HUSH-mediated silencing of intronless mobile elements" by Marta Seczynska et al., doi:10.1038/s41586-021-04228-1, Nature, 20 Jan 2022. "Genome surveillance by HUSH-mediated silencing of intronless mobile elements" by Marta Seczynska et al., doi:10.1038/s41586-021-04228-1, Nature, 20 Jan 2022.
 Molecular basis for the initiation of DNA primer synthesis by A.W.H. Li, K. Zabrady, L.J. Bainbridge, doi:10.1038/s41586-022-04695-0, Nature, 04 May 2022. Molecular basis for the initiation of DNA primer synthesis by A.W.H. Li, K. Zabrady, L.J. Bainbridge, doi:10.1038/s41586-022-04695-0, Nature, 04 May 2022.
 Break-induced replication: unraveling each step by Liping Liu and Anna Malkova, doi:10.1016/j.tig.2022.03.011, Trends in Genetics, 19 Apr 2022. Break-induced replication: unraveling each step by Liping Liu and Anna Malkova, doi:10.1016/j.tig.2022.03.011, Trends in Genetics, 19 Apr 2022.
 Overlapping genes in natural and engineered genomes by B.W. Wright, M.P. Molloy and P.R. Jaschke, doi:10.1038/s41576-021-00417-w, Nat Rev Genet, March 2022. Overlapping genes in natural and engineered genomes by B.W. Wright, M.P. Molloy and P.R. Jaschke, doi:10.1038/s41576-021-00417-w, Nat Rev Genet, March 2022.
 Gene-rich germline-restricted chromosomes in black-winged fungus gnats evolved through hybridization by Christina N Hodson et al., doi:10.1371/journal.pbio.3001559, PLoS Biol, 25 Feb 2022. Gene-rich germline-restricted chromosomes in black-winged fungus gnats evolved through hybridization by Christina N Hodson et al., doi:10.1371/journal.pbio.3001559, PLoS Biol, 25 Feb 2022.
 Taming transposable elements in vertebrates: from epigenetic silencing to domestication by Miguel Vasconcelos Almeida et al., doi:10.1016/j.tig.2022.02.009, Trends in Genetics, 17 Mar 2022. Taming transposable elements in vertebrates: from epigenetic silencing to domestication by Miguel Vasconcelos Almeida et al., doi:10.1016/j.tig.2022.02.009, Trends in Genetics, 17 Mar 2022.
 01 Mar 2022: ...a horizontal gene transfer created a new epigenetic system in animals.... 01 Mar 2022: ...a horizontal gene transfer created a new epigenetic system in animals....
 Throwing away DNA: programmed downsizing in somatic nuclei by Katherine H.I. Drotos et al., doi:10.1016/j.tig.2022.02.003, Trends in Genetics, 25 Feb 2022. Throwing away DNA: programmed downsizing in somatic nuclei by Katherine H.I. Drotos et al., doi:10.1016/j.tig.2022.02.003, Trends in Genetics, 25 Feb 2022.
 Essential Genes Protected from Mutation by Dan Robitzski, The Scientist, 25 Jan 2022; re: Essential Genes Protected from Mutation by Dan Robitzski, The Scientist, 25 Jan 2022; re:
 Mutation bias reflects natural selection in Arabidopsis thaliana by JG Monroe, T Srikant, P Carbonell-Bejerano et al., doi:10.1038/s41586-021-04269-6, Nature, 12 Jan 2022. Mutation bias reflects natural selection in Arabidopsis thaliana by JG Monroe, T Srikant, P Carbonell-Bejerano et al., doi:10.1038/s41586-021-04269-6, Nature, 12 Jan 2022.
 Overlapping genes in natural and engineered genomes by Bradley W. Wright et al., doi:10.1038/s41576-021-00417-w, Nat Rev Genet, 05 Oct 2021. Overlapping genes in natural and engineered genomes by Bradley W. Wright et al., doi:10.1038/s41576-021-00417-w, Nat Rev Genet, 05 Oct 2021.
 Long-range promoter-enhancer contacts are conserved during evolution and contribute to gene expression robustness by Alexandre Laverré et al., doi:10.1101/gr.275901.121, Genome Res., 20 Dec 2021. Long-range promoter-enhancer contacts are conserved during evolution and contribute to gene expression robustness by Alexandre Laverré et al., doi:10.1101/gr.275901.121, Genome Res., 20 Dec 2021.
 Reactivation of transposable elements following hybridization in fission yeast by Sergio Tusso et al., doi:10.1101/gr.276056.121, Genome Res., 14 Dec 2021. Reactivation of transposable elements following hybridization in fission yeast by Sergio Tusso et al., doi:10.1101/gr.276056.121, Genome Res., 14 Dec 2021.
 30 Jan 2022: Directed mutation in human regulatory genes 30 Jan 2022: Directed mutation in human regulatory genes
 Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? by Eduardo P. C. Rocha and David Bikard, doi:10.1371/journal.pbio.3001514, PLoS Biol, 13 Jan 2022. Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? by Eduardo P. C. Rocha and David Bikard, doi:10.1371/journal.pbio.3001514, PLoS Biol, 13 Jan 2022.
 20 Dec 2021: Chimeric RNAs become human genes?! 20 Dec 2021: Chimeric RNAs become human genes?!
"Coordinated spatiotemporal expression of large sets of genes is required for the development and homeostasis of organisms. To achieve this goal, organisms use myriad strategies where they form operons, utilize bidirectional promoters, cluster genes, share enhancers among genes by DNA looping, and form topologically associated domains and transcriptional condensates."
 Regulatory mechanisms ensuring coordinated expression of functionally related genes, by Oriana Q.H. Zinani et al., Trends in Genetics, Jan 2022. Regulatory mechanisms ensuring coordinated expression of functionally related genes, by Oriana Q.H. Zinani et al., Trends in Genetics, Jan 2022.
|
 ...A Complex Gene Family Involved in Epitranscriptomic Regulation and Other Epigenetic Processes by Juliet M Wong and Jose M Eirin-Lopez, Mol. Biol. & Evol., Dec 2021. ...A Complex Gene Family Involved in Epitranscriptomic Regulation and Other Epigenetic Processes by Juliet M Wong and Jose M Eirin-Lopez, Mol. Biol. & Evol., Dec 2021.
 A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure by Steve L. Bonilla et al., doi:10.1126/science.abe8526, Science, 18 Nov 2021. A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure by Steve L. Bonilla et al., doi:10.1126/science.abe8526, Science, 18 Nov 2021.
 The H2A.Z-nuclesome code in mammals: emerging functions by Yolanda Colino-Sanguino, Susan J. Clark and Fatima Valdes-Mora, doi:10.1016/j.tig.2021.10.003, Trends in Genetics, 23 Oct 2021. The H2A.Z-nuclesome code in mammals: emerging functions by Yolanda Colino-Sanguino, Susan J. Clark and Fatima Valdes-Mora, doi:10.1016/j.tig.2021.10.003, Trends in Genetics, 23 Oct 2021.
 Structural basis of human transcription-DNA repair coupling by Kokic, G., Wagner, F.R., Chernev, A. et al., doi:10.1038/s41586-021-03906-4, Nature, 15 Sep 2021. Structural basis of human transcription-DNA repair coupling by Kokic, G., Wagner, F.R., Chernev, A. et al., doi:10.1038/s41586-021-03906-4, Nature, 15 Sep 2021.
 Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements by Yijing Zhang et al., doi:10.1101/gr.275658.121, Genome Res., 09 Sep 2021. Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements by Yijing Zhang et al., doi:10.1101/gr.275658.121, Genome Res., 09 Sep 2021.
 ...DSB repair is fast and accurate ("RecA finds homologous DNA by reduced dimensionality search") by Wiktor, J., Gynnå, A.H., Leroy, P. et al., doi:10.1038/s41586-021-03877-6, Nature, 01 Sep 2021. ...DSB repair is fast and accurate ("RecA finds homologous DNA by reduced dimensionality search") by Wiktor, J., Gynnå, A.H., Leroy, P. et al., doi:10.1038/s41586-021-03877-6, Nature, 01 Sep 2021.
 ...novel protein interaction networks associated with sites of transcription initiation and termination by Desislava P Staneva et al., doi:10.1101/gr.275368.121, Genome Res., 18 Aug 2021. ...novel protein interaction networks associated with sites of transcription initiation and termination by Desislava P Staneva et al., doi:10.1101/gr.275368.121, Genome Res., 18 Aug 2021.
 31 Aug 2021: How could darwinian evolution invent programming for the whole ciliate MAC/MIC phenomenon: A discussion of a newly analysed gene-streamlining system in an unusual eukaryotic reproductive system. 31 Aug 2021: How could darwinian evolution invent programming for the whole ciliate MAC/MIC phenomenon: A discussion of a newly analysed gene-streamlining system in an unusual eukaryotic reproductive system.
 RNA editing restricts hyperactive ciliary kinases by Dongdong Li et al., doi:10.1126/science.abd8971, Science, 27 Aug 2021. RNA editing restricts hyperactive ciliary kinases by Dongdong Li et al., doi:10.1126/science.abd8971, Science, 27 Aug 2021.
 Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1 by Chen, Q., Plasencia, M., Li, Z. et al., Nature, 21 Jul 2021. ...how a small number of GRKs can selectively recognize and be activated by hundreds of different G-protein-coupled receptors. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1 by Chen, Q., Plasencia, M., Li, Z. et al., Nature, 21 Jul 2021. ...how a small number of GRKs can selectively recognize and be activated by hundreds of different G-protein-coupled receptors.
 Structural basis of human separase regulation by securin and CDK1-cyclin B1 by Yu, J., Raia, P., Ghent, C.M. et al., Nature, 21 Jul 2021. Structures ...shed light on the regulation of chromosome separation during the cell cycle. Structural basis of human separase regulation by securin and CDK1-cyclin B1 by Yu, J., Raia, P., Ghent, C.M. et al., Nature, 21 Jul 2021. Structures ...shed light on the regulation of chromosome separation during the cell cycle.
 A base-pair view of interactions between genes and their enhancers by Anne van Schoonhoven and Ralph Stadhouders, Nature, 09
Jun 2021. ...how DNA-binding proteins called transcription factors control gene expression. A base-pair view of interactions between genes and their enhancers by Anne van Schoonhoven and Ralph Stadhouders, Nature, 09
Jun 2021. ...how DNA-binding proteins called transcription factors control gene expression.
 Preparing macrophages for the future by Nagarajan Nandagopal et al., Science, 18 Jun 2021. ...some TFs ...not only control how genes respond in the present but also reconfigure the cell to control gene expression in response to future stimulation. Preparing macrophages for the future by Nagarajan Nandagopal et al., Science, 18 Jun 2021. ...some TFs ...not only control how genes respond in the present but also reconfigure the cell to control gene expression in response to future stimulation.
 Transposons Increase Transcriptional Complexity: The Good Parasite? by Joachim M. Surm and Yehu Moran, doi:10.1016/j.tig.2021.03.009, TiG, 12 Apr 2021. Transposons Increase Transcriptional Complexity: The Good Parasite? by Joachim M. Surm and Yehu Moran, doi:10.1016/j.tig.2021.03.009, TiG, 12 Apr 2021.
 On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life by Mathieu E. Rebeaud et al., doi:10.1073/pnas.2020885118, PNAS, 25 May 2021. On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life by Mathieu E. Rebeaud et al., doi:10.1073/pnas.2020885118, PNAS, 25 May 2021.
 Archaeal chromatin 'slinkies' are inherently dynamic complexes by Samuel Bowerman et al., doi:10.7554/eLife.65587, eLife, 02 Mar 2021. Archaeal chromatin 'slinkies' are inherently dynamic complexes by Samuel Bowerman et al., doi:10.7554/eLife.65587, eLife, 02 Mar 2021.
 Reorganization of the 3D chromatin architecture of rice genomes during heat stress by Zhe Liang et al., doi:10.1186/s12915-021-00996-4, BMC Biol, 19 Mar 2021. Our data uncovered higher-order chromatin architecture as a new layer in understanding transcriptional regulation in response to heat stress in rice. Reorganization of the 3D chromatin architecture of rice genomes during heat stress by Zhe Liang et al., doi:10.1186/s12915-021-00996-4, BMC Biol, 19 Mar 2021. Our data uncovered higher-order chromatin architecture as a new layer in understanding transcriptional regulation in response to heat stress in rice.
 Genomic Architecture of Rapid Parallel Adaptation to Fresh Water in a Wild Fish by Shao-Bing Zong, Yu-Long Li and Jin-Xian Liu, doi:10.1093/molbev/msaa290, Molecular Biology and Evolution, online 04 Nov 2020. The availability of beneficial standing genetic variation, large optimum shift between marine and freshwater habitats, and high efficiency of selection with large population size could lead to the observed rapid parallel adaptive genomic change. Genomic Architecture of Rapid Parallel Adaptation to Fresh Water in a Wild Fish by Shao-Bing Zong, Yu-Long Li and Jin-Xian Liu, doi:10.1093/molbev/msaa290, Molecular Biology and Evolution, online 04 Nov 2020. The availability of beneficial standing genetic variation, large optimum shift between marine and freshwater habitats, and high efficiency of selection with large population size could lead to the observed rapid parallel adaptive genomic change.
 QSER1 protects DNA methylation valleys from de novo methylation by Gary Dixon et al., doi:10.1126/science.abd0875, Science, 09 Apr 2021. QSER1 protects DNA methylation valleys from de novo methylation by Gary Dixon et al., doi:10.1126/science.abd0875, Science, 09 Apr 2021.
 Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring by Kyu-Hyeon Yeom et al., doi:10.1101/gr.273904.120, Genome Res., online 08 Apr 2021. Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring by Kyu-Hyeon Yeom et al., doi:10.1101/gr.273904.120, Genome Res., online 08 Apr 2021.
 Local nucleosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair by Vinay Tripuraneni et al., doi:10.1101/gr.271155.120, Genome Res., 02 Apr 2021. This ...suggests that a DNA replication-independent mechanism exists to preserve epigenome organization following DSB repair. Local nucleosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair by Vinay Tripuraneni et al., doi:10.1101/gr.271155.120, Genome Res., 02 Apr 2021. This ...suggests that a DNA replication-independent mechanism exists to preserve epigenome organization following DSB repair.
 Nonhomologous end joining: new accessory factors fine tune the machinery by Dipayan Ghosh and Sathees C. Raghavan, doi:10.1016/j.tig.2021.03.001, TiG, 27 Mar 2021. Nonhomologous end joining: new accessory factors fine tune the machinery by Dipayan Ghosh and Sathees C. Raghavan, doi:10.1016/j.tig.2021.03.001, TiG, 27 Mar 2021.
 Anything but Ordinary - Emerging Splicing Mechanisms in Eukaryotic Gene Regulation by Niels H. Gehring and Jean-Yves Roignant, doi:10.1016/j.tig.2020.10.008, TiG, 01 Apr 2021. Are there other noncanonical splicing events that remain to be identified? Anything but Ordinary - Emerging Splicing Mechanisms in Eukaryotic Gene Regulation by Niels H. Gehring and Jean-Yves Roignant, doi:10.1016/j.tig.2020.10.008, TiG, 01 Apr 2021. Are there other noncanonical splicing events that remain to be identified?
 Deciphering cis-regulatory grammar with deep learning by Emily R. Miraldi et al., doi:10.1038/s41588-021-00814-1, Nat Genet, 08 Mar 2021. Deciphering cis-regulatory grammar with deep learning by Emily R. Miraldi et al., doi:10.1038/s41588-021-00814-1, Nat Genet, 08 Mar 2021.
 RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination by Sijie Liu, Yu Hua, Jingna Wang et al., doi:10.1016/j.cell.2021.01.048, Cell, 23 Feb 2021. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination by Sijie Liu, Yu Hua, Jingna Wang et al., doi:10.1016/j.cell.2021.01.048, Cell, 23 Feb 2021.
 Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure by Jan Zrimec et al., doi:10.1038/s41467-020-19921-4, Nat Commun, 01 Dec 2020; and commentary:
How genetic motifs conduct "the music of life", Chalmers University of Technology, +Newswise, 28 Jan 2021. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure by Jan Zrimec et al., doi:10.1038/s41467-020-19921-4, Nat Commun, 01 Dec 2020; and commentary:
How genetic motifs conduct "the music of life", Chalmers University of Technology, +Newswise, 28 Jan 2021.
 Evidence supporting a viral origin of the eukaryotic nucleus by Philip J.L. Bell, doi:10.1016/j.virusres.2020.198168, Virus Research, Nov 2020. The eukaryotic system to uncouple transcription from translation is complex and employs hundreds of genes that act in concert. Evidence supporting a viral origin of the eukaryotic nucleus by Philip J.L. Bell, doi:10.1016/j.virusres.2020.198168, Virus Research, Nov 2020. The eukaryotic system to uncouple transcription from translation is complex and employs hundreds of genes that act in concert.
 A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores by Pablo Lara-Gonzalez et al., doi:10.1126/science.abc1424, Science, 01 Jan 2021. ...the detailed molecular choreography that allows a single, unattached kinetochore to arrest cell division. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores by Pablo Lara-Gonzalez et al., doi:10.1126/science.abc1424, Science, 01 Jan 2021. ...the detailed molecular choreography that allows a single, unattached kinetochore to arrest cell division.
 Histone variants in archaea and the evolution of combinatorial chromatin complexity by Kathryn M. Stevens et al., doi:10.1073/pnas.2007056117, PNAS, 29 Dec 2020. ...the ancestor of eukaryotes might have already had complex chromatin. Histone variants in archaea and the evolution of combinatorial chromatin complexity by Kathryn M. Stevens et al., doi:10.1073/pnas.2007056117, PNAS, 29 Dec 2020. ...the ancestor of eukaryotes might have already had complex chromatin.
 Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path by Thomas Dodd, Margherita Botto et al., Nature Communications, 23 Oct 2020. Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path by Thomas Dodd, Margherita Botto et al., Nature Communications, 23 Oct 2020.
 Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes by Liewei L. Yan and Hani S. Zaher, doi:10.1016/j.molcel.2020.11.033, Molecular Cell, 17 Dec 2020. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes by Liewei L. Yan and Hani S. Zaher, doi:10.1016/j.molcel.2020.11.033, Molecular Cell, 17 Dec 2020.
 Exo1 recruits Cdc5 polo kinase to MutLγ to ensure efficient meiotic crossover formation by Aurore Sanchez et al, doi:10.1073/pnas.2013012117, PNAS, 16 Nov 2020. Exo1 recruits Cdc5 polo kinase to MutLγ to ensure efficient meiotic crossover formation by Aurore Sanchez et al, doi:10.1073/pnas.2013012117, PNAS, 16 Nov 2020.
 We simply cannot go on being so vague about 'function' by W. Ford Doolittle, Genome Biol, 18 Dec 2018. We simply cannot go on being so vague about 'function' by W. Ford Doolittle, Genome Biol, 18 Dec 2018.
 Deep conservation of the enhancer regulatory code in animals by Emily S. Wong et al., doi:10.1126/science.aax8137, Science, 06 Nov 2020. Our results suggest the existence of an ancient and conserved, yet flexible, genomic regulatory syntax that has been repeatedly co-opted into cell type-specific gene regulatory networks across the animal kingdom. Deep conservation of the enhancer regulatory code in animals by Emily S. Wong et al., doi:10.1126/science.aax8137, Science, 06 Nov 2020. Our results suggest the existence of an ancient and conserved, yet flexible, genomic regulatory syntax that has been repeatedly co-opted into cell type-specific gene regulatory networks across the animal kingdom.
 90S pre-ribosome transformation into the primordial 40S subunit by Jingdong Cheng et al., doi:10.1126/science.abb4119, Science, 18 Sep 2020. 90S pre-ribosome transformation into the primordial 40S subunit by Jingdong Cheng et al., doi:10.1126/science.abb4119, Science, 18 Sep 2020.
 Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin by Silvija Bilokapic et al., doi:10.1038/s41586-020-2725-7, Nature, 16 Sep 2020. Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin by Silvija Bilokapic et al., doi:10.1038/s41586-020-2725-7, Nature, 16 Sep 2020.
 Massive project reveals complexity of gene regulation by Elizabeth Pennisi, Science, 11 Sep 2020. Massive project reveals complexity of gene regulation by Elizabeth Pennisi, Science, 11 Sep 2020.
 Expanded ENCODE delivers invaluable genomic encyclopedia by Chung-Chau Hon and Piero Carninci, Nature, 29 Jul 2020. Expanded ENCODE delivers invaluable genomic encyclopedia by Chung-Chau Hon and Piero Carninci, Nature, 29 Jul 2020.
 Principles of Epigenetic Homeostasis Shared Between Flowering Plants and Mammals by Ben P. Williams and Mary Gehring, doi:10.1016/j.tig.2020.06.019, Trends in Genetics, 2020. Maintaining epigenetic states through mitotic or meiotic cell divisions ...requires continual reestablishment of epigenetic information by enzymatic writers and erasers. Principles of Epigenetic Homeostasis Shared Between Flowering Plants and Mammals by Ben P. Williams and Mary Gehring, doi:10.1016/j.tig.2020.06.019, Trends in Genetics, 2020. Maintaining epigenetic states through mitotic or meiotic cell divisions ...requires continual reestablishment of epigenetic information by enzymatic writers and erasers.
 Dynamic human MutSα MutLα complexes compact mismatched DNA by Kira C. Bradford et al., doi:10.1073/pnas.1918519117, PNAS, 14 Jul 2020; and Dynamic human MutSα MutLα complexes compact mismatched DNA by Kira C. Bradford et al., doi:10.1073/pnas.1918519117, PNAS, 14 Jul 2020; and
 Recurrent mismatch binding by MutS mobile clamps on DNA localizes repair complexes nearby by Pengyu Hao et al., doi:10.1073/pnas.1918517117, PNAS, 15 Jul 2020; and commentary:
Genome Guardians Stop and Reel in DNA to Correct Replication Errors by Tracey Peake, NC State News (+Newswise), 16 Jul 2020. Recurrent mismatch binding by MutS mobile clamps on DNA localizes repair complexes nearby by Pengyu Hao et al., doi:10.1073/pnas.1918517117, PNAS, 15 Jul 2020; and commentary:
Genome Guardians Stop and Reel in DNA to Correct Replication Errors by Tracey Peake, NC State News (+Newswise), 16 Jul 2020.
 Contact area dependent cell communication and the morphological invariance of ascidian embryogenesis by Lèo Guignard1, Ulla-Maj Fiúza et al., doi:10.1126/science.aar5663, Science, 10 Jul 2020. Contact area dependent cell communication and the morphological invariance of ascidian embryogenesis by Lèo Guignard1, Ulla-Maj Fiúza et al., doi:10.1126/science.aar5663, Science, 10 Jul 2020.
 Hybrid Gene Origination Creates Human-Virus Chimeric Proteins during Infection by Jessica Sook Yuin Ho, Matthew Angel, Yixuan Ma, Elizabeth Sloan et al., doi:10.1016/j.cell.2020.05.035, Cell, 25 Jun 2020. ...a mechanism employed by sNSVs [segmented negative strand RNA viruses] to generate chimeric host-virus genes. Hybrid Gene Origination Creates Human-Virus Chimeric Proteins during Infection by Jessica Sook Yuin Ho, Matthew Angel, Yixuan Ma, Elizabeth Sloan et al., doi:10.1016/j.cell.2020.05.035, Cell, 25 Jun 2020. ...a mechanism employed by sNSVs [segmented negative strand RNA viruses] to generate chimeric host-virus genes.
 Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria by Jerónimo Rodr guez-Beltrán et al., doi:10.1073/pnas.2001240117, PNAS, 22 Jun 2020. Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria by Jerónimo Rodr guez-Beltrán et al., doi:10.1073/pnas.2001240117, PNAS, 22 Jun 2020.
 Adaptive evolution among cytoplasmic piRNA proteins leads to decreased genomic auto-immunity by Luyang Wang, Daniel A. Barbash and Erin S. Kelleher, doi:10.1371/journal.pgen.1008861, PLoS Genet, 11 Jun 2020. Adaptive evolution among cytoplasmic piRNA proteins leads to decreased genomic auto-immunity by Luyang Wang, Daniel A. Barbash and Erin S. Kelleher, doi:10.1371/journal.pgen.1008861, PLoS Genet, 11 Jun 2020.
 Multilayered mechanisms ensure that short chromosomes recombine in meiosis by Murakami, H., Lam, I., Huang, P. et al., doi:10.1038/s41586-020-2248-2, Nature, 04 Jun 2020. Multilayered mechanisms ensure that short chromosomes recombine in meiosis by Murakami, H., Lam, I., Huang, P. et al., doi:10.1038/s41586-020-2248-2, Nature, 04 Jun 2020.
 Global fitness landscapes of the Shine-Dalgarno sequence by Syue-Ting Kuo, Ruey-Lin Jahn,Yuan-Ju Cheng et al., doi:10.1101/gr.260182.119, Genome Res., 18 May 2020. A mechanism for "directed mutation?" ...the genotype-fitness correlation of SD promotes its evolvability by steadily supplying beneficial mutations across fitness landscapes.... Global fitness landscapes of the Shine-Dalgarno sequence by Syue-Ting Kuo, Ruey-Lin Jahn,Yuan-Ju Cheng et al., doi:10.1101/gr.260182.119, Genome Res., 18 May 2020. A mechanism for "directed mutation?" ...the genotype-fitness correlation of SD promotes its evolvability by steadily supplying beneficial mutations across fitness landscapes....
 21 May 2020 and 21 May 2020 and  20 May 2020: projects that observe robust software management. 20 May 2020: projects that observe robust software management.
 The TERB1-TERB2-MAJIN complex of mouse meiotic telomeres dates back to the common ancestor of metazoans by Irene da Cruz et al., doi:10.1186/s12862-020-01612-9, BMC Evol Biol, 14 May 2020. The TERB1-TERB2-MAJIN complex of mouse meiotic telomeres dates back to the common ancestor of metazoans by Irene da Cruz et al., doi:10.1186/s12862-020-01612-9, BMC Evol Biol, 14 May 2020.
 The Ccr4-Not complex monitors the translating ribosome for codon optimality by Robert Buschauer et al., doi:10.1126/science.aay6912, Science, 17 Apr 2020. The Ccr4-Not complex monitors the translating ribosome for codon optimality by Robert Buschauer et al., doi:10.1126/science.aay6912, Science, 17 Apr 2020.
 Repair, Removal, and Shutdown: It All Hinges on RNA Polymerase II Ubiquitylation by Kook Son and Orlando D. Schärer, doi:10.1016/j.cell.2020.02.053, Cell, 19 Mar 2020. Repair, Removal, and Shutdown: It All Hinges on RNA Polymerase II Ubiquitylation by Kook Son and Orlando D. Schärer, doi:10.1016/j.cell.2020.02.053, Cell, 19 Mar 2020.
 Caenorhabditis elegansADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons by Sylvia E. J. Fischer and Gary Ruvkun, doi:10.1073/pnas.1919028117, PNAS, 02 Mar 2020. Caenorhabditis elegansADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons by Sylvia E. J. Fischer and Gary Ruvkun, doi:10.1073/pnas.1919028117, PNAS, 02 Mar 2020.
 Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals by Greg Slodkowicz and Nick Goldman, doi:10.1073/pnas.1916786117, PNAS, 02 Mar 2020. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals by Greg Slodkowicz and Nick Goldman, doi:10.1073/pnas.1916786117, PNAS, 02 Mar 2020.
 Silencers, Enhancers, and the Multifunctional Regulatory Genome by Marc S. Halfon et al., doi:10.1016/j.tig.2019.12.005, Trends in Genetics, Mar 2020. Silencers, Enhancers, and the Multifunctional Regulatory Genome by Marc S. Halfon et al., doi:10.1016/j.tig.2019.12.005, Trends in Genetics, Mar 2020.
 Evolutionary History of GLISGenes Illuminates Their Roles in Cell Reprograming and Ciliogenesis by Yuuri Yasuoka et al., doi:10.1093/molbev/msz205, Molecular Biology and Evolution, 05 Sep 2019. Evolutionary History of GLISGenes Illuminates Their Roles in Cell Reprograming and Ciliogenesis by Yuuri Yasuoka et al., doi:10.1093/molbev/msz205, Molecular Biology and Evolution, 05 Sep 2019.
 Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates by Bo Xia et al., doi:10.1016/j.cell.2019.12.015, Cell, 23 Jan 2020; and commentary:
Scanning system in sperm may control rate of human evolution, NYU Langone Health via PhysOrg.com, 23 Jan 2020.
...transcription-coupled repair (TCR)... replaces faulty DNA patches just before transcription.... Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates by Bo Xia et al., doi:10.1016/j.cell.2019.12.015, Cell, 23 Jan 2020; and commentary:
Scanning system in sperm may control rate of human evolution, NYU Langone Health via PhysOrg.com, 23 Jan 2020.
...transcription-coupled repair (TCR)... replaces faulty DNA patches just before transcription....
 A kinase-dependent checkpoint prevents escape of immature ribosomes into the translating pool by Melissa D. Parker et al., doi:10.1371/journal.pbio.3000329, PLoS Biol, 13 Dec 2019. A kinase-dependent checkpoint prevents escape of immature ribosomes into the translating pool by Melissa D. Parker et al., doi:10.1371/journal.pbio.3000329, PLoS Biol, 13 Dec 2019.
 A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes by Joshua D. Eaton et al., doi:10.1101/gad.332833.119, Genes & Dev., 05 Dec 2019. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes by Joshua D. Eaton et al., doi:10.1101/gad.332833.119, Genes & Dev., 05 Dec 2019.
 Origin and Evolution of Two Independently Duplicated Genes ...in Caenorhabditisand Vertebrates by Diego A. Caraballo et al., G3, 03 Dec 2019. In both lineages, the catalytic domain of the duplicated genes was subjected to a strong purifying selective pressure, while the recognition domain was subjected to episodic positive diversifying selection. Origin and Evolution of Two Independently Duplicated Genes ...in Caenorhabditisand Vertebrates by Diego A. Caraballo et al., G3, 03 Dec 2019. In both lineages, the catalytic domain of the duplicated genes was subjected to a strong purifying selective pressure, while the recognition domain was subjected to episodic positive diversifying selection.
 Accessibility of promoter DNA is not the primary determinant of chromatin-mediated gene regulation by Răzvan V. Chereji, Peter R. Eriksson, Josefina Ocampo et al., Genome Res., Dec 2019. ...suggesting that transcription factors can penetrate heterochromatin. Accessibility of promoter DNA is not the primary determinant of chromatin-mediated gene regulation by Răzvan V. Chereji, Peter R. Eriksson, Josefina Ocampo et al., Genome Res., Dec 2019. ...suggesting that transcription factors can penetrate heterochromatin.
 Genomic sites hypersensitive to ultraviolet radiation by Sanjay Premi et al., PNAS, 26 Nov 2019. ...over 2,000 such genomic sites that are up to 170-fold more sensitive than the average site. These sites occur at specific locations near genes, so may let UV radiation drive direct changes in cell physiology rather than act through rare mutations. Genomic sites hypersensitive to ultraviolet radiation by Sanjay Premi et al., PNAS, 26 Nov 2019. ...over 2,000 such genomic sites that are up to 170-fold more sensitive than the average site. These sites occur at specific locations near genes, so may let UV radiation drive direct changes in cell physiology rather than act through rare mutations.
 14 Oct 2019: ...a group of yeasts ...capturing multiple genes from bacteria through horizontal gene transfer (HGT). 14 Oct 2019: ...a group of yeasts ...capturing multiple genes from bacteria through horizontal gene transfer (HGT).
 The nucleosome core particle remembers its position through DNA replication and RNA transcription by Gavin Schlissel and Jasper Rine, PNAS, 08 Oct 2019. The nucleosome core particle remembers its position through DNA replication and RNA transcription by Gavin Schlissel and Jasper Rine, PNAS, 08 Oct 2019.
 Cracking How 'Water Bears' Survive the Extremes, UC San Diego via newswise, 27 Sep 2019. ...Dsup protects cells by forming a protective cloud that shields DNA from hydroxyl radicals, which are produced by X-rays. Cracking How 'Water Bears' Survive the Extremes, UC San Diego via newswise, 27 Sep 2019. ...Dsup protects cells by forming a protective cloud that shields DNA from hydroxyl radicals, which are produced by X-rays.
 Widespread cis-regulatory convergence between the extinct Tasmanian tiger and gray wolf by Charles Y. Feigin et al., Genome Res., 18 Sep 2019. Our findings support the hypothesis that ...positive selection on cis-regulatory elements is likely to be an essential driver of adaptive convergent evolution.... Widespread cis-regulatory convergence between the extinct Tasmanian tiger and gray wolf by Charles Y. Feigin et al., Genome Res., 18 Sep 2019. Our findings support the hypothesis that ...positive selection on cis-regulatory elements is likely to be an essential driver of adaptive convergent evolution....
 05 Sep 2019: ...how a genome can seemingly intentionally respond to stress and pass those favorable adaptations on to its young. 05 Sep 2019: ...how a genome can seemingly intentionally respond to stress and pass those favorable adaptations on to its young.
 Evolution of intron splicing towards optimized gene expression is based on various Cis- and Trans-molecular mechanisms by Idan Frumkin et al., PLoS Biol, 23 Aug 2019. ...our work reveals various mechanistic pathways toward optimizations of intron splicing to ultimately adapt gene expression patterns to novel demands. Evolution of intron splicing towards optimized gene expression is based on various Cis- and Trans-molecular mechanisms by Idan Frumkin et al., PLoS Biol, 23 Aug 2019. ...our work reveals various mechanistic pathways toward optimizations of intron splicing to ultimately adapt gene expression patterns to novel demands.
 27 Aug 2019: Directed mutation can produce convergent evolution. 27 Aug 2019: Directed mutation can produce convergent evolution.
 Mammalian Mediator as a Functional Link between Enhancers and Promoters by Julie Soutourina, Cell, 22 Aug 2019. Mammalian Mediator as a Functional Link between Enhancers and Promoters by Julie Soutourina, Cell, 22 Aug 2019.
 The Unexpected Noncatalytic Roles of Histone Modifiers in Development and Disease by Yann Aubert et al., doi:10.1016/j.tig.2019.06.004, Trends in Genetics, 10 Jul 2019. The Unexpected Noncatalytic Roles of Histone Modifiers in Development and Disease by Yann Aubert et al., doi:10.1016/j.tig.2019.06.004, Trends in Genetics, 10 Jul 2019.
 10 Jul 2019: ...a large insertion of DNA from the endosymbiont bacteria Wolbachia ...integrated itself directly into the pillbug chromosome.... 10 Jul 2019: ...a large insertion of DNA from the endosymbiont bacteria Wolbachia ...integrated itself directly into the pillbug chromosome....
 Building bridges to move recombination complexes by Emeline Dubois et al., doi:10.1073/pnas.1901237116, PNAS, online 30 May 2019. ...the conundrum of how recombination complexes move from on-axis localization at coalignment to between-axis localization on SC central regions.... Building bridges to move recombination complexes by Emeline Dubois et al., doi:10.1073/pnas.1901237116, PNAS, online 30 May 2019. ...the conundrum of how recombination complexes move from on-axis localization at coalignment to between-axis localization on SC central regions....
 11 May 2019: beetles acquire and optimize GH45 gene 11 May 2019: beetles acquire and optimize GH45 gene
 Homolog Dependent Repair Following Dicentric Chromosome Breakage in Drosophila melanogaster by Jayaram Bhandari et al., Genetics, online 03 May 2019. Homolog Dependent Repair Following Dicentric Chromosome Breakage in Drosophila melanogaster by Jayaram Bhandari et al., Genetics, online 03 May 2019.
 Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes by Colin Thomas et al., PNAS, 26 Apr 2019. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes by Colin Thomas et al., PNAS, 26 Apr 2019.
 Genetic paradox explained by nonsense by Miles F. Wilkinson, Nature, 11 Apr 2019. ...the upregulation of compensatory genes is specifically triggered by mutations that generate short nucleotide sequences known as premature termination codons (PTCs). These sequences – also known as nonsense codons – signal the early cessation of the translation of messenger RNAs into proteins. Genetic paradox explained by nonsense by Miles F. Wilkinson, Nature, 11 Apr 2019. ...the upregulation of compensatory genes is specifically triggered by mutations that generate short nucleotide sequences known as premature termination codons (PTCs). These sequences – also known as nonsense codons – signal the early cessation of the translation of messenger RNAs into proteins.
 18 Apr 2019: the "grammar" of proteins can be investigated using tools borrowed from linguistics – Lijia Yu et al. 18 Apr 2019: the "grammar" of proteins can be investigated using tools borrowed from linguistics – Lijia Yu et al.
 08 Apr 2019: Helpful genetic mutations can be induced by environmental stress... (our comments about): 08 Apr 2019: Helpful genetic mutations can be induced by environmental stress... (our comments about):
 What is mutation? A chapter in the series: How microbes "jeopardize" the modern synthesis by Devon M. Fitzgerald and Susan M. Rosenberg, PLoS Genet., 01 Apr 2019. These mechanisms reveal a picture of highly regulated mutagenesis, up-regulated temporally by stress responses and activated when cells/organisms are maladapted to their environments—when stressed—potentially accelerating adaptation. Mutation is also nonrandom in genomic space, with multiple simultaneous mutations falling in local clusters, which may allow concerted evolution—the multiple changes needed to adapt protein functions and protein machines encoded by linked genes. What is mutation? A chapter in the series: How microbes "jeopardize" the modern synthesis by Devon M. Fitzgerald and Susan M. Rosenberg, PLoS Genet., 01 Apr 2019. These mechanisms reveal a picture of highly regulated mutagenesis, up-regulated temporally by stress responses and activated when cells/organisms are maladapted to their environments—when stressed—potentially accelerating adaptation. Mutation is also nonrandom in genomic space, with multiple simultaneous mutations falling in local clusters, which may allow concerted evolution—the multiple changes needed to adapt protein functions and protein machines encoded by linked genes.
 Transposable elements drive rapid phenotypic variation in Capsella rubella by Xiao-Min Niu et al., PNAS, online 15 Mar 2019. These results indicate that TE insertions drive rapid phenotypic variation, which could potentially help adapting to novel environments in species with limited genetic variation. Transposable elements drive rapid phenotypic variation in Capsella rubella by Xiao-Min Niu et al., PNAS, online 15 Mar 2019. These results indicate that TE insertions drive rapid phenotypic variation, which could potentially help adapting to novel environments in species with limited genetic variation.
 A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line by Bing Yang, Xia Xu, Logan Russell, et al., PLoS Genet., online 22 Feb 2019. A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line by Bing Yang, Xia Xu, Logan Russell, et al., PLoS Genet., online 22 Feb 2019.
 Evolution of resilience in protein interactomes across the tree of life by Marinka Zitnik et al., PNAS, online 14 Feb 2019. ...organisms stave off collapse through all manner of backup and workaround mechanisms.... Evolution of resilience in protein interactomes across the tree of life by Marinka Zitnik et al., PNAS, online 14 Feb 2019. ...organisms stave off collapse through all manner of backup and workaround mechanisms....
 Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments by Stephanie Fanucchi et al., v 51, Nature Genetics, online 10 Dec 2018. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments by Stephanie Fanucchi et al., v 51, Nature Genetics, online 10 Dec 2018.
 Nucleosome Positioning by an Evolutionarily Conserved Chromatin Remodeler Prevents Aberrant DNA Methylation in Neurospora by Andrew D. Klocko et al., doi:10.1534/genetics.118.301711, Genetics, online 15 Dec 2018. Nucleosome Positioning by an Evolutionarily Conserved Chromatin Remodeler Prevents Aberrant DNA Methylation in Neurospora by Andrew D. Klocko et al., doi:10.1534/genetics.118.301711, Genetics, online 15 Dec 2018.
 A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication by Monica S. Guo, Diane L. Haakonsen et al., doi:10.1016/j.cell.2018.08.029, Cell, 04 Oct 2018. A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication by Monica S. Guo, Diane L. Haakonsen et al., doi:10.1016/j.cell.2018.08.029, Cell, 04 Oct 2018.
 10 Oct 2018: Elegant and precise genetic programs guide the forces that allow seemingly identical starting cells to develop into highly specialized entities.... 10 Oct 2018: Elegant and precise genetic programs guide the forces that allow seemingly identical starting cells to develop into highly specialized entities....
 The NORAD lncRNA assembles a topoisomerase complex critical for genome stability by Mathias Munschauer et al., doi:10.1038/s41586-018-0453-z, Nature, 27 Aug 2018. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability by Mathias Munschauer et al., doi:10.1038/s41586-018-0453-z, Nature, 27 Aug 2018.
 The organization of genome duplication is a critical determinant of the landscape of genome maintenance by Blanca Gómez-Escoda and Pei-Yun Jenny Wu, doi:10.1101/gr.224527.117, Genome Res., 22 Jun 2018. The organization of genome duplication is a critical determinant of the landscape of genome maintenance by Blanca Gómez-Escoda and Pei-Yun Jenny Wu, doi:10.1101/gr.224527.117, Genome Res., 22 Jun 2018.
 RES complex is associated with intron definition and required for zebrafish early embryogenesis byJuan Pablo Fernandez, Miguel Angel Moreno-Mateos et al., PLoS Genet., 03 Jul 2018. ...long introns surrounding short exons are recognized and spliced through "exon definition" mechanisms.... RES complex is associated with intron definition and required for zebrafish early embryogenesis byJuan Pablo Fernandez, Miguel Angel Moreno-Mateos et al., PLoS Genet., 03 Jul 2018. ...long introns surrounding short exons are recognized and spliced through "exon definition" mechanisms....
 Uncovering universal rules governing the selectivity of the archetypal DNA glycosylase TDG by Thomas Dodd et al., PNAS, online 21 May 2018. Our results show that DNA sculpting, dynamic glycosylase interactions, and stabilizing contacts collectively provide a powerful mechanism for the detection and discrimination of modified bases and epigenetic marks in DNA. Uncovering universal rules governing the selectivity of the archetypal DNA glycosylase TDG by Thomas Dodd et al., PNAS, online 21 May 2018. Our results show that DNA sculpting, dynamic glycosylase interactions, and stabilizing contacts collectively provide a powerful mechanism for the detection and discrimination of modified bases and epigenetic marks in DNA.
 Dynamic Architecture of DNA Repair Complexes and the Synaptonemal Complex at Sites of Meiotic Recombination by Alexander Woglar and Anne M. Villeneuve, Cell, doi:10.1016/j.cell.2018.03.066, online 10 May 2018. Meiotic double-strand breaks (DSBs) are generated and repaired in a highly regulated manner to ensure formation of crossovers (COs) while also enabling efficient non-CO repair to restore genome integrity. Dynamic Architecture of DNA Repair Complexes and the Synaptonemal Complex at Sites of Meiotic Recombination by Alexander Woglar and Anne M. Villeneuve, Cell, doi:10.1016/j.cell.2018.03.066, online 10 May 2018. Meiotic double-strand breaks (DSBs) are generated and repaired in a highly regulated manner to ensure formation of crossovers (COs) while also enabling efficient non-CO repair to restore genome integrity.
 A Post-Transcriptional Feedback Mechanism for Noise Suppression and Fate Stabilization by Maike M.K. Hansen, Winnie Y. Wen, Elena Ingerman et al., Cell, doi:10.1016/j.cell.2018.04.005, online 10 May 2018. A Post-Transcriptional Feedback Mechanism for Noise Suppression and Fate Stabilization by Maike M.K. Hansen, Winnie Y. Wen, Elena Ingerman et al., Cell, doi:10.1016/j.cell.2018.04.005, online 10 May 2018.
 Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock by Vinesh Vinayachandran et al., Genome Res., online 14 Feb 2018. Together, these findings reveal protein genome interactions that are robustly reprogrammed in precise and uniform ways far beyond what is elicited by changes in gene expression. Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock by Vinesh Vinayachandran et al., Genome Res., online 14 Feb 2018. Together, these findings reveal protein genome interactions that are robustly reprogrammed in precise and uniform ways far beyond what is elicited by changes in gene expression.
 Stable Intronic Sequence RNAs Engage in Feedback Loops by Jun Wei Pek, Trends in Genetics, online 01 Feb 2018. The use of sisRNAs as mediators for local feedback control may be a general phenomenon. Stable Intronic Sequence RNAs Engage in Feedback Loops by Jun Wei Pek, Trends in Genetics, online 01 Feb 2018. The use of sisRNAs as mediators for local feedback control may be a general phenomenon.
 Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock by Vinesh Vinayachandran et al., Genome Res., online 14 Feb 2018. Our findings reveal a precise positional organization of proteins bound at most genes, some of which rapidly reorganize within minutes of heat shock. Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock by Vinesh Vinayachandran et al., Genome Res., online 14 Feb 2018. Our findings reveal a precise positional organization of proteins bound at most genes, some of which rapidly reorganize within minutes of heat shock.
 RNA Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates by William H. Palmer et al., doi:10.1534/genetics.117.300567, Genetics, 01 Feb 2018. RNA Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates by William H. Palmer et al., doi:10.1534/genetics.117.300567, Genetics, 01 Feb 2018.
 Protecting and Diversifying the Germline by Ryan J. Gleason et al., doi:10.1534/genetics.117.300208, Genetics, 01 Feb 2018. Protecting and Diversifying the Germline by Ryan J. Gleason et al., doi:10.1534/genetics.117.300208, Genetics, 01 Feb 2018.
 Systematic discovery of antiphage defense systems in the microbial pangenome by Shany Doron, Sarah Melamed et al., doi:10.1126/science.aar4120, Science, 25 Jan 2018. Our data also suggest a common, ancient ancestry of innate immunity components shared between animals, plants, and bacteria. Systematic discovery of antiphage defense systems in the microbial pangenome by Shany Doron, Sarah Melamed et al., doi:10.1126/science.aar4120, Science, 25 Jan 2018. Our data also suggest a common, ancient ancestry of innate immunity components shared between animals, plants, and bacteria.
 DNA mismatch repair preferentially protects genes from mutation by Eric J. Belfield, Zhong Jie Ding et al., doi:10.1101/gr.219303.116, Genome Res., 12 Dec 2017. DNA mismatch repair preferentially protects genes from mutation by Eric J. Belfield, Zhong Jie Ding et al., doi:10.1101/gr.219303.116, Genome Res., 12 Dec 2017.
 Structural basis for the initiation of eukaryotic transcription-coupled DNA repair by Jun Xu et al., Nature, 30 Nov 2017. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair by Jun Xu et al., Nature, 30 Nov 2017.
 Mismatch repair prefers exons by Dashiell J Massey and Amnon Koren, Nature Genetics, Dec 2017. Mismatch repair prefers exons by Dashiell J Massey and Amnon Koren, Nature Genetics, Dec 2017.
 Rapid Gene Family Evolution of a Nematode Sperm Protein Despite Sequence Hyper-conservation by Katja R. Kasimatis and Patrick C. Phillips, G3, online 21 Nov 2017. Rapid Gene Family Evolution of a Nematode Sperm Protein Despite Sequence Hyper-conservation by Katja R. Kasimatis and Patrick C. Phillips, G3, online 21 Nov 2017.
 Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae by Rui Bai, Chuangye Yan, Ruixue Wan et al., Cell, 16 Nov 2017. Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae by Rui Bai, Chuangye Yan, Ruixue Wan et al., Cell, 16 Nov 2017.
 The Mobile World of Transposable Elements by Caryn Navarro, Trends in Genetics, Nov 2017. The Mobile World of Transposable Elements by Caryn Navarro, Trends in Genetics, Nov 2017.
 23 Aug 2017: ...a different code embedded in histone marks.... 23 Aug 2017: ...a different code embedded in histone marks....
 15 Jul 2017: Several studies have suggested that TE [transposable element] insertions have contributed to the rewiring and evolution of regulatory networks by recruiting multiple genes into the same regulatory circuit. 15 Jul 2017: Several studies have suggested that TE [transposable element] insertions have contributed to the rewiring and evolution of regulatory networks by recruiting multiple genes into the same regulatory circuit.
 06 Jul 2017: How bacteria remember and defend against harmful viruses. 06 Jul 2017: How bacteria remember and defend against harmful viruses.
 22 May 2017: ...ERVL LTRs provide molecular mechanisms for stochastically scanning, rewiring, and recycling genetic information on an extraordinary scale. 22 May 2017: ...ERVL LTRs provide molecular mechanisms for stochastically scanning, rewiring, and recycling genetic information on an extraordinary scale.
 Conflict Resolution in the Genome: How Transcription and Replication Make It Work by Stephan Hamper and Karlene A. Cimprich, doi:10.1016/j.cell.2016.09.053, Cell, 01 Dec 2016. Conflicts between the transcription and replication complexes represent a potent source of genome instability in both prokaryotes and eukaryotes, and cells have evolved multiple transcription- and replication-dependent mechanisms to control, minimize, and overcome such encounters. Conflict Resolution in the Genome: How Transcription and Replication Make It Work by Stephan Hamper and Karlene A. Cimprich, doi:10.1016/j.cell.2016.09.053, Cell, 01 Dec 2016. Conflicts between the transcription and replication complexes represent a potent source of genome instability in both prokaryotes and eukaryotes, and cells have evolved multiple transcription- and replication-dependent mechanisms to control, minimize, and overcome such encounters.
 24 Jul 2016: A cell's deciphering arsenal.... 24 Jul 2016: A cell's deciphering arsenal....
 14 Jun 2016: Robust software management systems must effect the assembly, deployment, repair and optimization of acquired genetic programs.... 14 Jun 2016: Robust software management systems must effect the assembly, deployment, repair and optimization of acquired genetic programs....
 Cis-regulatory architecture of a brain signaling center predates the origin of chordates by Yao Yao et al., doi:10.1038/ng.3542, Nature Genetics, online 11 Apr 2016. Cis-regulatory architecture of a brain signaling center predates the origin of chordates by Yao Yao et al., doi:10.1038/ng.3542, Nature Genetics, online 11 Apr 2016.
 07 Apr 2016: ...molecular-resolution reconstruction of a central assembly of the human spliceosome. 07 Apr 2016: ...molecular-resolution reconstruction of a central assembly of the human spliceosome.
 28 Apr 2015: Diversity-generating retroelements (DGRs) use mutagenic reverse transcription and retrohoming to generate myriad variants of a target gene. 28 Apr 2015: Diversity-generating retroelements (DGRs) use mutagenic reverse transcription and retrohoming to generate myriad variants of a target gene.
 30 Jan 2015: I guess we owe the evolution of pregnancy to what are effectively genomic parasites. 30 Jan 2015: I guess we owe the evolution of pregnancy to what are effectively genomic parasites.
 19 Jan 2015: ...Deliberate killing of nonimmune cells ...releases DNA and makes it accessible for HGT. 19 Jan 2015: ...Deliberate killing of nonimmune cells ...releases DNA and makes it accessible for HGT.
 07 July 2014: ...not only may mutations be non-random but horizontal gene transfer too need not be random. 07 July 2014: ...not only may mutations be non-random but horizontal gene transfer too need not be random.
 20 Dec 2012: Evolution: A View from the 21st Century by James A. Shapiro 20 Dec 2012: Evolution: A View from the 21st Century by James A. Shapiro
 Quantifying the mechanisms of domain gain in animal proteins by Marija Buljan, Adam Frankish and Alex Bateman, doi:10.1186/gb-2010-11-7-r74; and commentary: Quantifying the mechanisms of domain gain in animal proteins by Marija Buljan, Adam Frankish and Alex Bateman, doi:10.1186/gb-2010-11-7-r74; and commentary:
 How do proteins gain new domains? by Joseph A Marsh and Sarah A Teichmann, doi:10.1186/gb-2010-11-7-126, Genome Biology, 15 Jul 2010. How do proteins gain new domains? by Joseph A Marsh and Sarah A Teichmann, doi:10.1186/gb-2010-11-7-126, Genome Biology, 15 Jul 2010.
 Enard D, Depaulis F, Crollius HR, "Human and Non-Human Primate Genomes Share Hotspots of Positive Selection" [link], PLoS Genet 6(2): e1000840. doi:10.1371/journal.pgen.1000840, online 5 Feb 2010. "Our results show that positive selection affecting the same genes independently in human and other primates is a common phenomenon and is not restricted to specific functions such as defence against pathogens or reproduction." Enard D, Depaulis F, Crollius HR, "Human and Non-Human Primate Genomes Share Hotspots of Positive Selection" [link], PLoS Genet 6(2): e1000840. doi:10.1371/journal.pgen.1000840, online 5 Feb 2010. "Our results show that positive selection affecting the same genes independently in human and other primates is a common phenomenon and is not restricted to specific functions such as defence against pathogens or reproduction."
 16 Jan 2010: The chimpanzee and human Y chromosomes have undergone wholesale renovation since the species diverged.... 16 Jan 2010: The chimpanzee and human Y chromosomes have undergone wholesale renovation since the species diverged....
 9 May 2006: The structure of a bacterial enzyme that inserts mobile gene cassettes has been resolved by French biochemists and geneticists. 9 May 2006: The structure of a bacterial enzyme that inserts mobile gene cassettes has been resolved by French biochemists and geneticists.
 24 Feb 2006: Retroposed genes have contributed to human evolution. 24 Feb 2006: Retroposed genes have contributed to human evolution.
 6 Aug 2005: Parallel evolution has been observed in fruitflies. 6 Aug 2005: Parallel evolution has been observed in fruitflies.
 24 Mar 2005: Plants can overwrite unhealthy genes. 24 Mar 2005: Plants can overwrite unhealthy genes.
 28 Feb 2005: Can pre-existing genetic programs be pieced together? 28 Feb 2005: Can pre-existing genetic programs be pieced together?
 29 Oct 2004: Pack-MULE transposable elements mediate gene evolution in plants. 29 Oct 2004: Pack-MULE transposable elements mediate gene evolution in plants.
 31 Dec 2003: Stress can increase the rate of horizontal gene transfer. 31 Dec 2003: Stress can increase the rate of horizontal gene transfer.
 30 Jun 2003: Introns can cause new stretches of DNA to be precisely inserted into genomes. 30 Jun 2003: Introns can cause new stretches of DNA to be precisely inserted into genomes.
 27 Sep 2000: Prions can turn on genetic programs. 27 Sep 2000: Prions can turn on genetic programs.
 03 Nov 1998: Two geneticists find evidence for "a predominating integration mechanism," that inserts acquired foreign genes into genomes in clustered fragments. 03 Nov 1998: Two geneticists find evidence for "a predominating integration mechanism," that inserts acquired foreign genes into genomes in clustered fragments.
References
1. Michael T. Madigan, John M. Martinko and Jack Parker, Brock Biology of Microorganisms, 8th ed., 1997. p 97.
1.5. Michael J. Daly and Kenneth W. Minton, Resistance to Radiation, Science, 24 November 1995.
2. Bruce Alberts et al., The Molecular Biology of the Cell, 3rd ed., 1994. p 268.
2.5. James D. Watson et al., The Molecular Biology of the Gene, 4th ed., 1987. p 485.
2.6. Miroslav Radman. Enzymes of evolutionary change, doi:10.1038/44738, Nature, 28 October 1999.
3. Shozo Yokoyama et al.,
Adaptive evolution of color vision of the Comoran coelacanth..., PNAS, 25 May 1999.
3.5. Susumu Ohno, Evolution by Gene Duplication, Springer-Verlag Publishing Company, 1970. p 55.
4. Eva C. M. Nowack et al., Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora, PNAS, online 10 Oct 2016.
4.5. Blair G. Paul et al., Targeted diversity generation by intraterrestrial archaea and archaeal viruses, doi:10.1038/ncomms7585, n 6585 v 6, Nature Communications, 23 Mar 2015.
5. Gary M. Dunny, The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution, doi:10.1098/rstb.2007.2043, Phil. Trans. R. Soc. B, 29 Jul 2007.
6. Emily S. Wong et al., Deep conservation of the enhancer regulatory code in animals, Science, 06 Nov 2020; and commentary: Enhancers: Conserved in Activity, Not in Sequence by Jack Lee, The Scientist, 01 Nov 2021.
Related CA Webpages
 What Is Life? includes the suggestion, A Cell Is Like a Computer. What Is Life? includes the suggestion, A Cell Is Like a Computer.
 Why Sexual Reproduction? includes a section titled Gene Conversion. Why Sexual Reproduction? includes a section titled Gene Conversion.
 Viruses and Other Gene Transfer Mechanisms is relevant. Viruses and Other Gene Transfer Mechanisms is relevant.
 How is it Possible? has some earlier refences for adaptive and directed mutation. How is it Possible? has some earlier refences for adaptive and directed mutation.
![]()
|


