July 26
 SETI looks at panspermia. [Thanks, Marsbugs.]
SETI looks at panspermia. [Thanks, Marsbugs.]
 Panspermia: Spreading Life Through the Universe, by Seth Shostak, SETI Institute, 24 July 2003.
Panspermia: Spreading Life Through the Universe, by Seth Shostak, SETI Institute, 24 July 2003.
 Introduction... is a related CA webpage.
Introduction... is a related CA webpage.
 An Atmospheric Test of Cometary Panspermia is a related CA webpage.
An Atmospheric Test of Cometary Panspermia is a related CA webpage.
July 20
More genes seem to precede the need for themselves. A team at the University of Wisconsin found that unicellular choanoflagellates express members of protein families previously found only in multicelled animals:
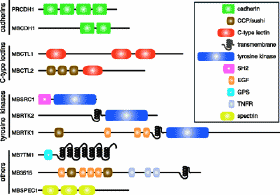 "In marked contrast to their simple lifestyle, choanoflagellates express members of a wide variety of protein families involved in animal cell interactions, including cadherins, C-type lectins, tyrosine kinases, and a G protein-coupled receptor, as well as several multidomain polypeptides that contain protein-protein interaction domains involved in signaling and adhesion in animals [such as the epidermal growth factor motif, Src homology 2 domain, tumor necrosis factor receptor domain, and sushi or complement control protein domain]."
"In marked contrast to their simple lifestyle, choanoflagellates express members of a wide variety of protein families involved in animal cell interactions, including cadherins, C-type lectins, tyrosine kinases, and a G protein-coupled receptor, as well as several multidomain polypeptides that contain protein-protein interaction domains involved in signaling and adhesion in animals [such as the epidermal growth factor motif, Src homology 2 domain, tumor necrosis factor receptor domain, and sushi or complement control protein domain]."
King et al. suspect more. They write, "We have sampled just a fraction of the choanoflagellate proteome. The diversity of choanoflagellate proteins predicted to function in cell interactions suggests that additional proteins shared exclusively with animals will be discovered through sequencing the entire choanoflagellate genome. ...It may then be possible to determine whether entire regulatory pathways linking receptor-based signaling inputs to gene regulation and cell behavior predate the origin of animals." As darwinists, they reason,
"The discovery of multiple signaling and adhesion gene family members in choanoflagellates demonstrates that key proteins required for animal development evolved before the origin of animals." If so, they wonder,
"The existence in unicellular choanoflagellates of proteins used for cell adhesion and signal transduction in animals prompts the question of their ancestral function in the progenitor of animals and choanoflagellates." You bet it does. Here they introduce a list of speculations.
Reconstructing evolution that happened hundreds of millions of years ago from clues in today's genomes requires substantial faith. However, if the method is valid, we think the evidence in this study supports cosmic ancestry, whereunder existing species acquire new genetic programs by gene transfer. Only after acquisition will the programs be expressed in new phenotypes. If, on the other hand, new programs are gradually composed by the darwinian method, the ones in this study must have originally served purposes different from the ones they serve in animals today. To account for this phenomenon darwinians must invent more stories.
 Nicole King et al., "Evolution of Key Cell Signaling and Adhesion Protein Families Predates Animal Origins" [abstract], p 361-363 v 301, Science, 18 Jul 2003.
Nicole King et al., "Evolution of Key Cell Signaling and Adhesion Protein Families Predates Animal Origins" [abstract], p 361-363 v 301, Science, 18 Jul 2003.
 Ancient ancestor's legacy of life, by David Whitehouse, BBC News, 22 July 2003.
Ancient ancestor's legacy of life, by David Whitehouse, BBC News, 22 July 2003.
 Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
 Metazoan Genes Older Than Metazoa? is a related CA webpage.
Metazoan Genes Older Than Metazoa? is a related CA webpage.
July 10
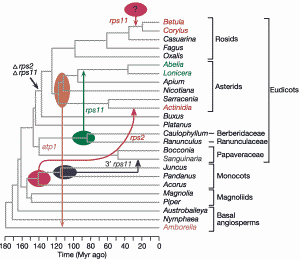 The role of gene transfer in evolution is greater than previously thought. A team of US biologists reached this conclusion after a study of transferred mitochondrial genes in plants. They write,
The role of gene transfer in evolution is greater than previously thought. A team of US biologists reached this conclusion after a study of transferred mitochondrial genes in plants. They write,
"Conventional genes are subject to evolutionarily frequent HGT during plant evolution and... plants can donate DNA horizontally to other plants."
"Eukaryotic genomes regularly acquire genes by means of horizontal events that are relatively recent, datable, and definable as to donor and recipient."
"We believe the five cases reported here are merely the tip of a large iceberg of mitochondrial HGT in plants."
"It seems likely that plant nuclear genomes are also significantly affected by HGT." And they wonder,
"Is HGT driven predominantly by potential vectoring agents such as viruses, bacteria, fungi, insects, pollen or even meteorites; or by the transformational uptake of plant DNA released into the soil; or by unrelated plants occasionally grafting together?" [Thanks, Hans-Peter Wheeler.]
 Ulfar Bergthorsson et al., "Widespread horizontal transfer of mitochondrial genes in flowering plants" [abstract], p 197-201 v 424, Nature, 10 July 2003.
Ulfar Bergthorsson et al., "Widespread horizontal transfer of mitochondrial genes in flowering plants" [abstract], p 197-201 v 424, Nature, 10 July 2003.
 CL Bishop, "Plant-to-plant horizontal gene transfer" [text], The Scientist, 10 July 2003.
CL Bishop, "Plant-to-plant horizontal gene transfer" [text], The Scientist, 10 July 2003.
 Plant genes imported from unrelated species more often than previously thought..., Indiana University, 7 July 2003.
Plant genes imported from unrelated species more often than previously thought..., Indiana University, 7 July 2003.
 Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
July 8
 NASA launches the second Mars Rover, nearly two weeks later than scheduled. This mission and its twin, launched last month, should land on opposite sides of Mars in January, 2004. The rovers will look for signs that the planet may have once supported life.
NASA launches the second Mars Rover, nearly two weeks later than scheduled. This mission and its twin, launched last month, should land on opposite sides of Mars in January, 2004. The rovers will look for signs that the planet may have once supported life.
 The World Goes to Mars, Astrobiology Magazine, 9 July 2003.
The World Goes to Mars, Astrobiology Magazine, 9 July 2003.
 NASA Launches Mars Rover After Delays, The Associated Press, ABCNews, 8 July 2003.
NASA Launches Mars Rover After Delays, The Associated Press, ABCNews, 8 July 2003.
 2nd Mars Rover Finally Is Bound for Red Planet, by Allison M. Heinrichs and Usha Lee McFarling, Los Angeles Times, 8 July 2003.
2nd Mars Rover Finally Is Bound for Red Planet, by Allison M. Heinrichs and Usha Lee McFarling, Los Angeles Times, 8 July 2003.
 Life on Mars! is the related CA webpage.
Life on Mars! is the related CA webpage.
June 30
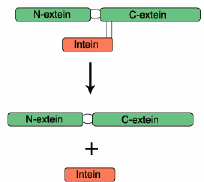 Introns can cause new stretches of DNA to be precisely inserted into genomes. While hints of this capability were already known, introns seem even more remarkable now, according to a review in Science. "Introns serve as templates for making other proteins. Among these are spectacular enzymes that inject new stretches of DNA into precisely defined spots in a genome." Tools like this one would be necessary for the integration of new genetic programs, as in cosmic ancestry.
Introns can cause new stretches of DNA to be precisely inserted into genomes. While hints of this capability were already known, introns seem even more remarkable now, according to a review in Science. "Introns serve as templates for making other proteins. Among these are spectacular enzymes that inject new stretches of DNA into precisely defined spots in a genome." Tools like this one would be necessary for the integration of new genetic programs, as in cosmic ancestry.The article is also informative about inteins [see figure], which are similar to introns by being sequences that are ulimately removed. But inteins are edited out of amino acid sequences (after translation), whereas introns are edited out of mRNA sequences (before translation).
 Ingred Wickelgren, "Spinning Junk Into Gold" [abstract], p 1646-1649 v 300, Science, 13 June 2003.
Ingred Wickelgren, "Spinning Junk Into Gold" [abstract], p 1646-1649 v 300, Science, 13 June 2003.
 Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
Viruses... is a related CA webpage about gene transfer. [Next-What'sNEW about HGT-Prev]
 Introns... is a related CA webpage.
Introns... is a related CA webpage.

June 24
An in-depth commentary on the Viking mission's tests for life on Mars is posted. "We have received biology data that we believe to be good data...," said NASA's Jim Martin at the 1:30 pm news briefing on 31 July 1976. And on 7 August, NASA's Norman Horowitz said, "There's a possibility that this is biological, [but] there are many other possibilities that have to be excluded." Arguments for nonbiological interpretations of the Viking data are clearer here than elsewhere. [Thanks, Marsbugs.]
 Mars: Life Pinned on Viking Horns? Astrobiology Magazine, 22 June 2003.
Mars: Life Pinned on Viking Horns? Astrobiology Magazine, 22 June 2003.
 Life on Mars! — a CA webpage with more about the Viking mission.
Life on Mars! — a CA webpage with more about the Viking mission.
June 21
A transcript of Thursday's Internet chat about about Cosmic Ancestry is available. [Thanks, moderator Micah Sparacio and all who participated.]
 Brig Klyce: Cosmic Ancestry - The Modern Version of Panspermia: Transcript of ICSD Internet chat, 9:00-10:00 PM EDT, 19 June 2003.
Brig Klyce: Cosmic Ancestry - The Modern Version of Panspermia: Transcript of ICSD Internet chat, 9:00-10:00 PM EDT, 19 June 2003.
 Introduction... is a related CA webpage.
Introduction... is a related CA webpage.
June 19
Horizontal gene transfer as a significant evolutionary driver may require an addendum to the Darwinian synthesis. So say commentators Raymond and Blankenship in PNAS about a study of 78 plastid-targeted proteins from a eukaryotic alga. It showed, "Even by conservative measures, ~21% of these genes have likely been acquired by HGT." Furthermore, "These horizontally transferred genes span a varied swath of functions...."
 "Most intriguingly, two of the genes from [Archibald et al.'s] analysis indicate HGT from different bacteria, significant not only as an example of prokaryote-to-eukaryote gene transfer but also because these acquired genes initially would have not had the proper leader sequence for import into the plastid. Whether the appropriate targeting sequence was incorporated de novo through gene conversion or some other mechanism of homologous or orthologous replacement is not clear, but this remarkable finding certainly invokes new ideas on how genes are assimilated into a genome." Finally, "It now seems clear that many organisms, eukaryotes and prokaryotes, are certainly able to mimic evolutionary jumps through HGT."
"Most intriguingly, two of the genes from [Archibald et al.'s] analysis indicate HGT from different bacteria, significant not only as an example of prokaryote-to-eukaryote gene transfer but also because these acquired genes initially would have not had the proper leader sequence for import into the plastid. Whether the appropriate targeting sequence was incorporated de novo through gene conversion or some other mechanism of homologous or orthologous replacement is not clear, but this remarkable finding certainly invokes new ideas on how genes are assimilated into a genome." Finally, "It now seems clear that many organisms, eukaryotes and prokaryotes, are certainly able to mimic evolutionary jumps through HGT."
 J.M. Archibald, M.B. Rogers, M. Toop, K.-i Ishida and P.J. Keeling, "Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans" [abstract], p 7678–7683 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
J.M. Archibald, M.B. Rogers, M. Toop, K.-i Ishida and P.J. Keeling, "Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans" [abstract], p 7678–7683 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
 Jason Raymond and Robert E. Blankenship, "Horizontal gene transfer in eukaryotic algal evolution" [title page], p 7419-7420 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
Insertions and deletions, important for human evolution, occur at preferred sites. A team from UCSD used computer analysis of genome data to reach this conclusion. They elaborate, "...Mammalian genomes are mosaics of fragile regions with high propensity for rearrangements and solid regions with low propensity for rearrangements." [Thanks, Stan Franklin.]
Jason Raymond and Robert E. Blankenship, "Horizontal gene transfer in eukaryotic algal evolution" [title page], p 7419-7420 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
Insertions and deletions, important for human evolution, occur at preferred sites. A team from UCSD used computer analysis of genome data to reach this conclusion. They elaborate, "...Mammalian genomes are mosaics of fragile regions with high propensity for rearrangements and solid regions with low propensity for rearrangements." [Thanks, Stan Franklin.]

 UCSD Researchers Estimate Approximately 400 Fragile Regions in the Human Genome That Are Vulnerable to Evolutionary 'Earthquakes', UCSD News, 16 June 2003.
UCSD Researchers Estimate Approximately 400 Fragile Regions in the Human Genome That Are Vulnerable to Evolutionary 'Earthquakes', UCSD News, 16 June 2003.
 Pavel Pevzner and Glenn Tesler, "Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution" [abstract], p 7672-7677 v 100, Proc. Natl. Acad. Sci. USA, 24 Jun 2003.
The evolutionary importance of insertions and deletions gets more notice from an international team who compared orthologous sequences from the human and chimp genomes (1.87 and 1.75 million nucleotides, respectively). They conclude, "these data suggest that evolution may have used the mechanistically more drastic indels instead of the more subtle single-nucleotide substitutions for shaping the recently emerged primate species."
Pavel Pevzner and Glenn Tesler, "Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution" [abstract], p 7672-7677 v 100, Proc. Natl. Acad. Sci. USA, 24 Jun 2003.
The evolutionary importance of insertions and deletions gets more notice from an international team who compared orthologous sequences from the human and chimp genomes (1.87 and 1.75 million nucleotides, respectively). They conclude, "these data suggest that evolution may have used the mechanistically more drastic indels instead of the more subtle single-nucleotide substitutions for shaping the recently emerged primate species."
 Tatsuya Anzai et al., "Comparative sequencing of human and chimpanzee MHC class I regions unveils insertion/deletions as the major path to genomic divergence" [abstract], p 7708-7713 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
Tatsuya Anzai et al., "Comparative sequencing of human and chimpanzee MHC class I regions unveils insertion/deletions as the major path to genomic divergence" [abstract], p 7708-7713 v 100, Proc. Natl. Acad. Sci. USA, 24 June 2003.
 Human Genome Search... is a related CA webpage.
These recent developments are consistent with cosmic ancestry, we suggest. In the new theory, evolutionary advances are made possible when new genetic programs are acquired by horizontal gene transfer. This paradigm shift has already been accepted for prokaryotes; now evidence for it among eukaryotes is accumulating. Naturally, it requires the insertion of large blocks of code. Then, placing imported genes or gene fragments into designated reception areas would make them easier to retrieve later, and it would reduce the risk of damage to the receiving genome. How the genome would then assimilate the new software is not known, but the installation system would probably include mechanisms that are remarkable and intriguing to darwinists.
Human Genome Search... is a related CA webpage.
These recent developments are consistent with cosmic ancestry, we suggest. In the new theory, evolutionary advances are made possible when new genetic programs are acquired by horizontal gene transfer. This paradigm shift has already been accepted for prokaryotes; now evidence for it among eukaryotes is accumulating. Naturally, it requires the insertion of large blocks of code. Then, placing imported genes or gene fragments into designated reception areas would make them easier to retrieve later, and it would reduce the risk of damage to the receiving genome. How the genome would then assimilate the new software is not known, but the installation system would probably include mechanisms that are remarkable and intriguing to darwinists.
 Viruses... is a CA webpage about horizontal gene transfer. [Next-What'sNEW about HGT-Prev]
Viruses... is a CA webpage about horizontal gene transfer. [Next-What'sNEW about HGT-Prev]
 Robust Software Management, a new webpage, has comments and speculations about mechanisms for installing genetic programs.
Robust Software Management, a new webpage, has comments and speculations about mechanisms for installing genetic programs.
June 10
 A live online chat about panspermia with Brig Klyce, this website's author, will take place at 9PM Eastern on Thursday, 19 June 2003 (if all the software works!) Visit the link below and download an applet to participate in or to watch the discussion. The chat is sponsored by The International Society for Complexity, Information, and Design, a creationist-oriented group. We welcome this opportunity to discuss panspermia.
A live online chat about panspermia with Brig Klyce, this website's author, will take place at 9PM Eastern on Thursday, 19 June 2003 (if all the software works!) Visit the link below and download an applet to participate in or to watch the discussion. The chat is sponsored by The International Society for Complexity, Information, and Design, a creationist-oriented group. We welcome this opportunity to discuss panspermia.
 Live Moderated Chat Event to be held at 9PM EDT, 19 June 2003.
Live Moderated Chat Event to be held at 9PM EDT, 19 June 2003.
June 3
 European Mars Express space probe is on the way to Mars. Due to arrive there in late December, "This first European Space Agency probe to head for another planet will enter an orbit around Mars, from where it will perform detailed studies of the planet's surface, its subsurface structures and its atmosphere. It will also deploy Beagle 2, a small autonomous station which will land on the planet, studying its surface and looking for possible signs of life, past or present."
European Mars Express space probe is on the way to Mars. Due to arrive there in late December, "This first European Space Agency probe to head for another planet will enter an orbit around Mars, from where it will perform detailed studies of the planet's surface, its subsurface structures and its atmosphere. It will also deploy Beagle 2, a small autonomous station which will land on the planet, studying its surface and looking for possible signs of life, past or present."
 Mars Express en route for the Red Planet, European Space Agency, 2 June 2003.
Mars Express en route for the Red Planet, European Space Agency, 2 June 2003.
 Beagle Hunts for Mars, Astrobiology Magazine, 4 June 2003.
Beagle Hunts for Mars, Astrobiology Magazine, 4 June 2003.
 Mars Express all set for launch, by Damian Carrington, NewScientist.com, 2 June 2003.
Mars Express all set for launch, by Damian Carrington, NewScientist.com, 2 June 2003.
 Europe's first trip to Mars launched, by Tom Clarke, Nature Science Update, 9 June 2003.
Europe's first trip to Mars launched, by Tom Clarke, Nature Science Update, 9 June 2003.
 Life on Mars! is the related CA webpage.
Life on Mars! is the related CA webpage.
May 29