What'sNEW October–December 2009
 26 December 2009 26 December 2009
...The protein, named ZBED6, is previously unknown, specific for placental mammals, and derived from an exapted DNA transposon. This finding of an important, acquired gene comes from new research by a collaboration of 25 scientist from Sweden and Boston's Broad Institute. The protein is widely expressed, binds to about 2,500 sites, and regulates many important steps in development and growth. The gene encoding it includes an open reading frame of approximately 2,900 base pairs or more than 900 codons. Its conservation among placental mammals is close to 100%.
 Click for full figure Click for full figure |
The research report observes, "The difference between less complex eukaryotes like Caenorhabditis elegans and more complex eukaryotes, such as human, is related not to the number of protein-coding genes, but rather to the complexity of the gene regulatory networks. A large proportion of vertebrate genomes is composed of transposable elements, and their integration in the genome has contributed to the evolution of regulatory networks. The majority of these transposable elements are retrotransposons, but 5%-10% are derived from DNA transposons. In the initial analysis of the first human genome assembly, Lander et al. identified 47 human genes derived from transposable elements, as many as 43 of these are derived from DNA transposons."
"We found evidence for a nonfunctional ZBED6 sequence at the orthologous positions in the Platypus and opossum genomes, but these genomes did not contain an extended open reading frame for ZBED6.... This implies that the integration of ZBED6 happened before the divergence of the monotremes from the other mammals, but that the gene has been inactivated or lost in monotremes and marsupials." [Couldn't it have been acquired more than once?] "Thus, ZBED6 must have evolved its essential function in the time span after the split between marsupial and placental mammals, but before the radiation of different orders of placental mammals."
Two sequences of 54 amino acids each, in zinc finger domains (see figure extract: click for full figure), are perfectly conserved across 20 of 26 listed species including pig, mouse, human, dog, cat, rat, horse and guinea pig. Apparently the DNA sequence encoding this enzyme arrived via a transposon, already composed. So how ZBED6 "evolved its essential function" seems to overlook the obvious — the function was already there. In any case, there is no evidence that it was gradually composed by mutation-and-natural-selection; and encountering a complete function so specific among long-enough random DNA sequences is absurdly unlikely. We think transposon-supplied genes like ZEBD6 add weight to the case for cosmic ancestry, in which new genes must be acquired by some form of horizontal gene transfer (HGT).
 Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, et al., "ZBED6, a Novel Transcription Factor Derived from a Domesticated DNA Transposon Regulates IGF2 Expression and Muscle Growth" [article], PLoS Biol 7(12): e1000256. doi:10.1371/journal.pbio.1000256, online Dec 2009. Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, et al., "ZBED6, a Novel Transcription Factor Derived from a Domesticated DNA Transposon Regulates IGF2 Expression and Muscle Growth" [article], PLoS Biol 7(12): e1000256. doi:10.1371/journal.pbio.1000256, online Dec 2009.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage. Viruses and Other Gene Transfer Mechanisms is the main related local webpage.
What'sNEW about HGT  | |
 24 December 2009 24 December 2009
...Interstellar dust includes a substantial fraction of biomaterial in various stages of degradation. We have modeled such an ensemble ...in precise agreement with the interstellar extinction observations.
 Kani Rauf and Chandra Wickramasinghe, "Evidence for biodegradation products in the interstellar medium" [preprint.pdf], doi:10.1017/S1473550409990334, p29-34 v9, International Journal of Astrobiology, Jan 2010. Kani Rauf and Chandra Wickramasinghe, "Evidence for biodegradation products in the interstellar medium" [preprint.pdf], doi:10.1017/S1473550409990334, p29-34 v9, International Journal of Astrobiology, Jan 2010.
 Hoyle and Wickramasinghe's Analysis of Interstellar Dust is the main related local webpage. Hoyle and Wickramasinghe's Analysis of Interstellar Dust is the main related local webpage.
 Thanks, Google Alerts. Thanks, Google Alerts.
 16 December 2009 16 December 2009
NASA's David S. McKay sends additional information and links pertaining to the evidence for life on Mars:
 From David S. McKay, 11 Dec 2009. From David S. McKay, 11 Dec 2009.
 Ancient life remains the most plausible explanation... What'sNEW, 1 Dec 2009. Ancient life remains the most plausible explanation... What'sNEW, 1 Dec 2009.
 16 December 2009 16 December 2009
 The giant [Marseillevirus, pictured] adopts genes from the other organisms, including eukaryotes, bacteria and other viruses.... — Boyer, Yutin et al.
The giant [Marseillevirus, pictured] adopts genes from the other organisms, including eukaryotes, bacteria and other viruses.... — Boyer, Yutin et al.
We propose that amoebae are 'melting pots' of microbial evolution where diverse forms emerge, including giant viruses with complex gene repertoires of various origins — coauthor Didier Raoult, Université de la Mediterranée, Marseille, France
 Mickaël Boyer, Natalya Yutin et al., "Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms" [abstract], doi:10.1073/pnas.0911354106, Proc. Natl. Acad. Sci. USA, online 9 Dec 2009. Mickaël Boyer, Natalya Yutin et al., "Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms" [abstract], doi:10.1073/pnas.0911354106, Proc. Natl. Acad. Sci. USA, online 9 Dec 2009.
 New giant virus discovered, Katherine Bagley, The Scientist, 7 Dec 2009. New giant virus discovered, Katherine Bagley, The Scientist, 7 Dec 2009.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage. Viruses and Other Gene Transfer Mechanisms is the main related local webpage.
What'sNEW about HGT  | |
 Thanks, Stan Franklin. Thanks, Stan Franklin.
 13 December 2009 13 December 2009
Some microorganisms can survive interplanetary journeys cocooned inside rocks blasted off planets by comet and asteroid impacts. That rocky panspermia is an effective mechanism for spreading life within a planetary system — Paul Davies, Arizona State University
This comment accompanies news that Jeffrey Touchman of ASU's School of Life Sciences will undertake new studies concerning the evolution of photosynthesis. The project will include sequencing the genomes of extremophiles — good candidates for transferring life among planets. Previous analysis from ASU showed that photosynthesis apparently evolved by gene transfer; and related studies implicate viruses as likely transfer agents. Perhaps the new research will notice that panspermia and gene transfer can explain a lot about the evolution of photosynthesis, and evolution in general.

 'Extreme' genes sheds light on origins of photosynthesis by Margaret Coulombe, Arizona State University, 11 Dec 2009. 'Extreme' genes sheds light on origins of photosynthesis by Margaret Coulombe, Arizona State University, 11 Dec 2009.
 Searching for the Origins of Photosynthesis: Experts study extremophiles by Tudor Vieru, Softpedia, 12 Dec 2009. Searching for the Origins of Photosynthesis: Experts study extremophiles by Tudor Vieru, Softpedia, 12 Dec 2009.
 Photosynthesis evolved by gene transfer: a related What'sNEW article, 24 Nov 2002. Photosynthesis evolved by gene transfer: a related What'sNEW article, 24 Nov 2002.
 Photosynthesis genes in a virus: a related What'sNEW article, 14 Aug 2003. Photosynthesis genes in a virus: a related What'sNEW article, 14 Aug 2003.
 More about photosynthesis by gene transfer: a related What'sNEW article, 27 Jul 2004. More about photosynthesis by gene transfer: a related What'sNEW article, 27 Jul 2004.
 Viruses... has more. Search for "photosynthesis". Viruses... has more. Search for "photosynthesis".
What'sNEW about HGT  | |
 Thanks, Google Alerts. Thanks, Google Alerts.
 11 December 2009 11 December 2009
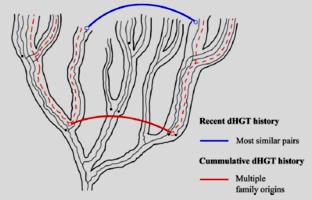 In recent years, the idea has grown that horizontal transfer is the dominant force in expanding the gene repertoires of bacterial genomes. In fact, an analysis of gene phylogenies in γ-proteobacteria suggests that only a small percentage of the genes found in extant species descend from the most recent common ancestor of this group. This idea implies that microbial genomes are highly dynamic entities, constantly acquiring and losing genes. Thus... for most organisms, the percentage of genes transferred at any point in the species' history could be close to 100%, independent of size. — Otto X. Cordero and Paulien Hogeweg, Theoretical Biology and Bioinformatics, University of Utrecht, The Netherlands.
In recent years, the idea has grown that horizontal transfer is the dominant force in expanding the gene repertoires of bacterial genomes. In fact, an analysis of gene phylogenies in γ-proteobacteria suggests that only a small percentage of the genes found in extant species descend from the most recent common ancestor of this group. This idea implies that microbial genomes are highly dynamic entities, constantly acquiring and losing genes. Thus... for most organisms, the percentage of genes transferred at any point in the species' history could be close to 100%, independent of size. — Otto X. Cordero and Paulien Hogeweg, Theoretical Biology and Bioinformatics, University of Utrecht, The Netherlands. This observation casually appears in a study that measures ancient and recent horizontal gene transfer (HGT) among bacteria. It caught our attention, to say the least.
 Otto X. Cordero and Paulien Hogeweg, "The impact of long-distance horizontal gene transfer on prokaryotic genome size" [abstract], doi:10.1073/pnas.0907584106, Proc. Natl. Acad. Sci. USA, online 9 Dec 2009. Otto X. Cordero and Paulien Hogeweg, "The impact of long-distance horizontal gene transfer on prokaryotic genome size" [abstract], doi:10.1073/pnas.0907584106, Proc. Natl. Acad. Sci. USA, online 9 Dec 2009.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage. Viruses and Other Gene Transfer Mechanisms is the main related local webpage.
What'sNEW about HGT  | |
 The Tree of Life: a related local webpage. The Tree of Life: a related local webpage.
 8 December 2009 8 December 2009
 Life may make the methane on Mars. That possibility was strengthened when one possible nonbiological source, ablation from meteorites, was ruled out at Imperial College London. Two specialists in astromaterials there simulated the meteoritic process and found that it could produce only a tiny fraction of the observed methane. Other nonbiolgical sources remain under consideration.
Life may make the methane on Mars. That possibility was strengthened when one possible nonbiological source, ablation from meteorites, was ruled out at Imperial College London. Two specialists in astromaterials there simulated the meteoritic process and found that it could produce only a tiny fraction of the observed methane. Other nonbiolgical sources remain under consideration.
 Richard W. Court and Mark A. Sephton, "Investigating the contribution of methane produced by ablating micrometeorites to the atmosphere of Mars" [abstract], doi:10.1016/j.epsl.2009.09.041, p382-385 v288, Earth and Planetary Science Letters, 15 Nov 2009. Richard W. Court and Mark A. Sephton, "Investigating the contribution of methane produced by ablating micrometeorites to the atmosphere of Mars" [abstract], doi:10.1016/j.epsl.2009.09.041, p382-385 v288, Earth and Planetary Science Letters, 15 Nov 2009.
 Life on Mars theory boosted by new methane study, EurekAlert!, 8 Dec 2009. Life on Mars theory boosted by new methane study, EurekAlert!, 8 Dec 2009.
 New Findings Say Mars Methane Comes from Life or Water - or Both by Nancy Atkinson, UniverseToday, 8 Dec 2009. New Findings Say Mars Methane Comes from Life or Water - or Both by Nancy Atkinson, UniverseToday, 8 Dec 2009.
 Life on Mars! is the main related local webpage. Search for "methane". Life on Mars! is the main related local webpage. Search for "methane".
 Thanks for an added link, Larry Klaes. Thanks for an added link, Larry Klaes.
 1 December 2009 1 December 2009
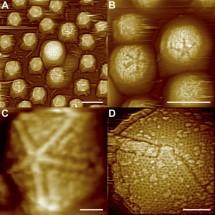
A meteorite designated ALH84001 became famous in 1996, when a group of scientists led by David McKay, Everett Gibson and Kathie Thomas-Keprta of NASA's Johnson Space Center announced that it contained evidence of past life on Mars. That claim has since been widely debated, and non-biological accounts of the same evidence have emerged.
 Now the NASA team has used new tools to model the principal non-biological scenarios; they find them untenable. "We conclude that the vast majority of the nanocrystal magnetites present in the carbonate disks [example, right] could not have formed by any of the currently proposed thermal decomposition scenarios." Therefore they believe, Ancient life remains the most plausible explanation for the materials and structures found in the Mars meteorite.
Now the NASA team has used new tools to model the principal non-biological scenarios; they find them untenable. "We conclude that the vast majority of the nanocrystal magnetites present in the carbonate disks [example, right] could not have formed by any of the currently proposed thermal decomposition scenarios." Therefore they believe, Ancient life remains the most plausible explanation for the materials and structures found in the Mars meteorite.
 K.L. Thomas-Keprta et al., "Origins of magnetite nanocrystals in Martian meteorite ALH84001" [abstract | 47-page PDF], doi:10.1016/j.gca.2009.05.064, p6631-6677 v73, Geochimica et Cosmochimica Acta, 1 Nov (online 16 Jun) 2009. K.L. Thomas-Keprta et al., "Origins of magnetite nanocrystals in Martian meteorite ALH84001" [abstract | 47-page PDF], doi:10.1016/j.gca.2009.05.064, p6631-6677 v73, Geochimica et Cosmochimica Acta, 1 Nov (online 16 Jun) 2009.
 New Study Adds to Finding of Ancient Life Signs In Mars Meteorite, NASA News Release J09-030, by William P. Jeffs, Johnson Space Center, Houston, 30 Nov 2009. New Study Adds to Finding of Ancient Life Signs In Mars Meteorite, NASA News Release J09-030, by William P. Jeffs, Johnson Space Center, Houston, 30 Nov 2009.
 NASA Technical Reports Server, with links to NASA abstracts of related papers. NASA Technical Reports Server, with links to NASA abstracts of related papers.
 Martian meteorite surrenders new secrets of possible life, by Craig Covault, Spaceflight Now, 24 Nov 2009. Martian meteorite surrenders new secrets of possible life, by Craig Covault, Spaceflight Now, 24 Nov 2009.
 Evidence of life on Mars lurks beneath surface of meteorite..., The Times (London), 27 Nov 2009. Evidence of life on Mars lurks beneath surface of meteorite..., The Times (London), 27 Nov 2009.
 New Study of Meteorite Provides More Evidence for Ancient Life on Mars, by Lisa Zyga, PhysOrg.com, 17 Dec 2009. New Study of Meteorite Provides More Evidence for Ancient Life on Mars, by Lisa Zyga, PhysOrg.com, 17 Dec 2009.
 Life on Mars! is the related local webpage, with a special section, What'sNEW: ALH84001. Life on Mars! is the related local webpage, with a special section, What'sNEW: ALH84001.
 Thanks Larry Klaes, Doron Goldberg, Jerry Chancellor, Google Alerts and Stan Franklin. Thanks Larry Klaes, Doron Goldberg, Jerry Chancellor, Google Alerts and Stan Franklin.
 28 November 2009 28 November 2009
There is no such a thing as a 'simple' bacterium — evolutionary biologists Howard Ochman and Rahul Raghavan
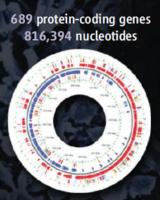 Mycoplasma pneumoniae is among the smallest bacteria that can be cultivated in vitro. Its circular genome (illustrated) measures 816,394 nucleotide pairs long, only a fifth as large as the genome of well-studied E. coli. A minimal free-living cell, M. pneumoniae was chosen by a partnership between the European Molecular Biology Laboratory in Heidelberg, Germany, and the Centro de Regulación Genómica in Barcelona, Spain, as the subject of extensive, quantitative studies. Three papers in Science report the results of these studies. One thing is clear: "Even the simplest of cells is more complex than expected."
Mycoplasma pneumoniae is among the smallest bacteria that can be cultivated in vitro. Its circular genome (illustrated) measures 816,394 nucleotide pairs long, only a fifth as large as the genome of well-studied E. coli. A minimal free-living cell, M. pneumoniae was chosen by a partnership between the European Molecular Biology Laboratory in Heidelberg, Germany, and the Centro de Regulación Genómica in Barcelona, Spain, as the subject of extensive, quantitative studies. Three papers in Science report the results of these studies. One thing is clear: "Even the simplest of cells is more complex than expected."
If any of the mainstream origin-of-life theories has merit, the simplest forms of life should be simple indeed. This coordinated study emphatically indicates otherwise. In invited commentary, Ochman and Raghaven suggest why.
 Howard Ochman and Rahul Raghavan, "Excavating the Functional Landscape of Bacterial Cells" [abstract], doi:10.1126/science.1183757, p1200-1201 v326, Science, 27 Nov 2009. Howard Ochman and Rahul Raghavan, "Excavating the Functional Landscape of Bacterial Cells" [abstract], doi:10.1126/science.1183757, p1200-1201 v326, Science, 27 Nov 2009.
 Sebastian Kühner, Vera van Noort et al., "Proteome Organization in a Genome-Reduced Bacterium" [abstract], doi:10.1126/science.1176343, p1235-1240 v326, Science, 27 Nov 2009. Sebastian Kühner, Vera van Noort et al., "Proteome Organization in a Genome-Reduced Bacterium" [abstract], doi:10.1126/science.1176343, p1235-1240 v326, Science, 27 Nov 2009.
 Eva Yus et al., "Impact of Genome Reduction on Bacterial Metabolism and Its Regulation" [abstract], doi:10.1126/science.1177263, p1263-1268 v326, Science, 27 Nov 2009. Eva Yus et al., "Impact of Genome Reduction on Bacterial Metabolism and Its Regulation" [abstract], doi:10.1126/science.1177263, p1263-1268 v326, Science, 27 Nov 2009.
 Marc Güell et al., "Transcriptome Complexity in a Genome-Reduced Bacterium" [abstract], doi:10.1126/science.1176951, p1268-1271 v326, Science, 27 Nov 2009. Marc Güell et al., "Transcriptome Complexity in a Genome-Reduced Bacterium" [abstract], doi:10.1126/science.1176951, p1268-1271 v326, Science, 27 Nov 2009.
 Lucas Laursen, "Single-celled life does a lot with very little" [html], doi:10.1038/news.2009.1110, Nature, online 26 Nov 2009. Lucas Laursen, "Single-celled life does a lot with very little" [html], doi:10.1038/news.2009.1110, Nature, online 26 Nov 2009.
 PDF: First-ever blueprint of a minimal cell is more complex than expected, The European Molecular Biology Laboratory (+ EurekAlert.org), 26 Nov 2009. PDF: First-ever blueprint of a minimal cell is more complex than expected, The European Molecular Biology Laboratory (+ EurekAlert.org), 26 Nov 2009.
 What Is Life? is a related local webpage, What Is Life? is a related local webpage,
 The RNA World is the main local webpage about origin-of-life theories. The RNA World is the main local webpage about origin-of-life theories.
 25 November 2009 25 November 2009
Butterflies and their larvae, caterpillars, were once separate species, brought together by hybridization, according to a controversial article published by Proceedings of the National Academy of Sciences. British biologist Donald Williamson's article containing this hypothesis was escorted through peer review by like-minded Academy member Lynn Margulis. After its online publication in August, opponents complained loudly; a vigorous rebuttal immediately follows it in the print issue. We are interested, because hybridization is a wholesale form of horizontal gene transfer (HGT).
A: Caterpillar anterior
B: Velvet worm
C: Cambrian fossil resembling a velvet worm © Williamson, PNAS, 2009
|
Williamson supports his theory mainly by noting morphological similarities between caterpillars and velvet worms, legged worms that were were apparently more widespread in the Cambrian period than today. Williamson also proposes tests for the hypothesis, one being genome size comparisons: hybrids should have more protein-coding genes than non-hybrids, he believes. The rebuttal by a pair of North American biologists claims that the relevant genome sizes are already known and do not sustain the theory. Besides, they find the whole thing ludicrous.
We are mildly amused by the tempest. If HGT is essential for macroevolutionary innovation, as we believe, some consequences of this paradigm shift are bound startle mainstream darwinists. This particular example may or may not be ultimately vindicated, but exploring it seems straightforward and harmless. Simply counting genes, however, is an over-simplified measurement that will not settle the issue. Whole genome sequences, and lots of them, are needed. Even with these, inference is required and conflicting interpretations can be defended. Of course, the best proof that HGT is not essential for macroevolutionary innovation would be demonstrations in quarantined systems.
Larval development is indeed interesting. In butterfly metamorphosis, the larval organs virtually dissolve and once-differentiated cells revert into stem cells again. These cells then develop into new organs and systems — amazing, really. Williamson and others have previously suggested that HGT could have introduced the genetic programs for this process, so the basic idea is not completely new. The outrage however, seems overzealous.
 Donald I. Williamson, "Caterpillars evolved from onychophorans by hybridogenesis" [abstract], doi:10.1073/pnas.0908357106, p19901-19905 v106, Proc. Natl. Acad. Sci. USA, 24 Nov (online 28 Aug) 2009. Donald I. Williamson, "Caterpillars evolved from onychophorans by hybridogenesis" [abstract], doi:10.1073/pnas.0908357106, p19901-19905 v106, Proc. Natl. Acad. Sci. USA, 24 Nov (online 28 Aug) 2009.
 Michael W. Hart and Richard K. Grosberg, "Caterpillars did not evolve from onychophorans by hybridogenesis" [abstract], doi:10.1073/pnas.0908357106, p19906-19909 v106, Proc. Natl. Acad. Sci. USA, 24 Nov 2009. Michael W. Hart and Richard K. Grosberg, "Caterpillars did not evolve from onychophorans by hybridogenesis" [abstract], doi:10.1073/pnas.0908357106, p19906-19909 v106, Proc. Natl. Acad. Sci. USA, 24 Nov 2009.
 Gonzalo Giribet, "On velvet worms and caterpillars: Science, fiction, or science fiction?" [extract], doi:10.1073/pnas.0910279106, E131 v106, Proc. Natl. Acad. Sci. USA, online only, 30 Oct 2009. Gonzalo Giribet, "On velvet worms and caterpillars: Science, fiction, or science fiction?" [extract], doi:10.1073/pnas.0910279106, E131 v106, Proc. Natl. Acad. Sci. USA, online only, 30 Oct 2009.
 Donald I. Williamson, "Reply to Giribet..." [extract], doi:10.1073/pnas.0912151106, E132 v106, Proc. Natl. Acad. Sci. USA, online only, 30 Oct 2009. Donald I. Williamson, "Reply to Giribet..." [extract], doi:10.1073/pnas.0912151106, E132 v106, Proc. Natl. Acad. Sci. USA, online only, 30 Oct 2009.
 Worst paper of the year?, Why Evolution Is True, by Jerry Coyne, 4 Sep 2009. Worst paper of the year?, Why Evolution Is True, by Jerry Coyne, 4 Sep 2009.
 National Academy as National Enquirer...? by Brendan Borrell, ScientificAmerican.com, 24 Aug 2009. National Academy as National Enquirer...? by Brendan Borrell, ScientificAmerican.com, 24 Aug 2009.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage. Viruses and Other Gene Transfer Mechanisms is the main related local webpage.
What'sNEW about HGT  | |
 Macroevolutionary Progress ...Without Gene Transfer? is a related local webpage. Macroevolutionary Progress ...Without Gene Transfer? is a related local webpage.
 Testing Darwinism versus Cosmic Ancestry: a related local webpage. Testing Darwinism versus Cosmic Ancestry: a related local webpage.
 Acquiring Genomes: our review of this book by Margulis and Sagan, posted 7 Jul 2002. Acquiring Genomes: our review of this book by Margulis and Sagan, posted 7 Jul 2002.
 Horizontal Gene Transfer: our book review mentioning the possible HGT of genes for larval forms, posted 4 Nov 1999. Horizontal Gene Transfer: our book review mentioning the possible HGT of genes for larval forms, posted 4 Nov 1999.
 22 November 2009 22 November 2009
 The deep sea is teeming with species that have never known sunlight, according to a news release from the Census of Marine Life, a systematic investigation of life on the ocean floor. The deepest ocean floor was once thought to be too barren, cold, dark and pressurized for life to flourish there. Wrong! Actually, "The abyssal fauna is so rich in species diversity and so poorly described that collecting a known species is an anomaly," says Dr. David Billett of UK's National Oceanography Centre.
The deep sea is teeming with species that have never known sunlight, according to a news release from the Census of Marine Life, a systematic investigation of life on the ocean floor. The deepest ocean floor was once thought to be too barren, cold, dark and pressurized for life to flourish there. Wrong! Actually, "The abyssal fauna is so rich in species diversity and so poorly described that collecting a known species is an anomaly," says Dr. David Billett of UK's National Oceanography Centre. Some of this life depends on food from higher levels that settles to the depths, but other species there survive entirely on inorganic chemicals. We suspect that species in the latter category could also survive in the deep ocean on Europa, for example.
 "From the Edge of Darkness to the Black Abyss: Marine Scientists Census 17,500+ Species and Counting" [PDF], Census of Marine Life, 22 Nov 2009. "From the Edge of Darkness to the Black Abyss: Marine Scientists Census 17,500+ Species and Counting" [PDF], Census of Marine Life, 22 Nov 2009.
 Census of Marine Life, the Office of Marine Programs, University of Rhode Island, Graduate School of Oceanography. Census of Marine Life, the Office of Marine Programs, University of Rhode Island, Graduate School of Oceanography.
 Life on Europa, Other Moons, Other Planets? is a local webpage with links about moons with ice-covered oceans. Life on Europa, Other Moons, Other Planets? is a local webpage with links about moons with ice-covered oceans.
 The extreme diversity and curious distribution of deep sea microbes...., another What'sNEW article about the Census of Marine Life, 19 Apr 2010. The extreme diversity and curious distribution of deep sea microbes...., another What'sNEW article about the Census of Marine Life, 19 Apr 2010.
 12 November 2009 12 November 2009
In this form the bacteria can survive for hundreds, perhaps millions, of years in a dormant state and, what's more, endure drought, extreme temperatures, radiation, and toxins that would quickly knock out unprotected bacteria. But spores read their surroundings, and as soon as conditions improve, they transform back into active bacteria. These words come from a press release about research by an interdisciplinary team from Lund University and the University of Connecticut concerning bacterial spores that can survive standard sterilization measures. Using a new technique, the team observes that the spores' hardiness "may be linked to dehydration-induced conformational changes in key enzymes." And they mention that the spores' dormancy and resistance have relevance for ...astrobiology!

 Erik P. Sunde et al., "The physical state of water in bacterial spores" [abstract], doi:10.1073/pnas.0908712106, p19334-19339 v106, Proc. Natl. Acad. Sci. USA, 17 Nov (online 5 Nov) 2009. Erik P. Sunde et al., "The physical state of water in bacterial spores" [abstract], doi:10.1073/pnas.0908712106, p19334-19339 v106, Proc. Natl. Acad. Sci. USA, 17 Nov (online 5 Nov) 2009.
 New explanation for nature's hardiest life form, EurekAlert, 12 Nov 2009. New explanation for nature's hardiest life form, EurekAlert, 12 Nov 2009.
 Bacteria: The Space Colonists: a related local webpage with more about hardy, long-surviving bacteria. Bacteria: The Space Colonists: a related local webpage with more about hardy, long-surviving bacteria.
 29 October 2009 29 October 2009
40,000 generations of E. Coli have been monitored in a long-term experiment that tracks cloned populations in quarantined environments. This experiment at Michigan State University exemplifies the surest way to measure the power of darwinian mutation-and-natural-selection in life. If the mainstream theory is correct, this power is virtually unlimited and new genetic programs should readily emerge.
The team managing the experiment reports that in one population, after 20,000 generations, 45 mutations were observed. Most of these, 29, were single-nucleotide polymorphisms; the remainder were deletions, insertions, or other polymorphisms. Relative fitness advantages (versus the ancestor) conferred by the mutations were rapid initially, but then slowed. Interestingly, all of the point mutations in coding regions were non-synonymous, and many were identical even in unrelated populations.
 Mutations and Relative fitness over 20,000 generations (except inset)
Mutations and Relative fitness over 20,000 generations (except inset)
Mutations: left scale | blue data — Circles show the total number of genomic changes in clones sampled at 2,500, 5,000, 10,000, 15,000 and 20,000 generations. The straight diagonal line represents a model where mutations accumulate uniformly over time. The thinner stepped curves define the 95% confidence interval for this linear model.
Relative fitness: green data | right scale — Squares show the level of this population's mean fitness relative to the ancestor, and the curve is a hyperbolic plus linear fit of this trajectory. Each fitness estimate is the mean of three assays; most of the spread of points around the fitness trajectory reflects statistical uncertainty inherent to the assays.
Inset: Mutations over 40,000 generations — The shallow, dash-dot segment shows the same mutations as the main (blue) graph through 20,000 generations. The steep, dotted final segment approximates the timecourse of genomic evolution after a mutator phenotype appeared by about generation 26,500, culminating in 653 mutations in the 40,000th-generation clone. Relative fitness is not shown. (Figure and caption adapted from Barrick, Yu et al., Nature 2009.)
|
Around generation 26,500, a phenotype emerged in the studied population whose mutation rate was roughly 20 times the previously observed rate (see inset graph). A single base-pair insertion that disabled the mutT gene was the apparent cause. The population subsequently fixed mostly neutral mutations and its genome shrank 1.2%.
The new report pays close attention to the discordance between the mutation rate (steady at first, then accelerating after about generation 26,500), and the relative fitness improvements (initially rapid, soon slowing, then flat when mutations accelerated.) In standard darwinian evolution, this discordance is surprising. But we believe that darwinian mutation-and-natural-selection can only toggle and optimize the genetic programs that are available. If so, fitness improvements in a closed system should slow and eventually stop, as may be apparent here. And genetic programs not initially present in a given system will not emerge unless they are supplied from elsewhere via some means of gene transfer. So we are not surprised that the latest report on this quarantined experiment mentions no new genetic programs. In sum, we think the experiment leaves room for doubt about the mainstream theory of evolution. We will keep watching.
| How is "fitness" measured? In this experiment, fitness is the relative ability of the subject population to outcompete, or outnumber, the others in a given environment. No substantial evolutionary innovation is required or implied. |
 Jeffrey E. Barrick, Dong Su Yu et al., "Genome evolution and adaptation in a long-term experiment with Escherichia coli" [abstract], doi:10.1038/nature08480, p 1243-1247 v 461, Nature, 29 Oct 2009. Jeffrey E. Barrick, Dong Su Yu et al., "Genome evolution and adaptation in a long-term experiment with Escherichia coli" [abstract], doi:10.1038/nature08480, p 1243-1247 v 461, Nature, 29 Oct 2009.
 Paul B. Rainey, "Evolutionary biology: Arrhythmia of tempo and mode" [html], doi:10.1038/4611219a, p 1219 v 461, Nature, 29 Oct 2009. Paul B. Rainey, "Evolutionary biology: Arrhythmia of tempo and mode" [html], doi:10.1038/4611219a, p 1219 v 461, Nature, 29 Oct 2009.
 Tanita Casci, "Evolution: The routes of adaptation" [html], doi:10.1038/nrg2706, p815 v10, Nature Reviews Genetics, Dec 2009. Tanita Casci, "Evolution: The routes of adaptation" [html], doi:10.1038/nrg2706, p815 v10, Nature Reviews Genetics, Dec 2009.
 ...Is Evolutionary Progress in a Closed System Possible? is a related webpage, with a special What'sNEW section, Lenski et al.. ...Is Evolutionary Progress in a Closed System Possible? is a related webpage, with a special What'sNEW section, Lenski et al..
 Macroevolutionary Progress ...Without Gene Transfer? is a related local webpage. Macroevolutionary Progress ...Without Gene Transfer? is a related local webpage.
 Testing Darwinism versus Cosmic Ancestry: a related local webpage. Testing Darwinism versus Cosmic Ancestry: a related local webpage.
 14 October 2009 14 October 2009
Instead of life originating here on Earth..., perhaps it came from somewhere else....

 Panspermia, by Jean Tate, Universe Today, 13 Oct 2009. Panspermia, by Jean Tate, Universe Today, 13 Oct 2009.
 Introduction: More Than Panspermia is a related local webpage. Introduction: More Than Panspermia is a related local webpage.
 Thanks, Google Alerts. Thanks, Google Alerts.
 9 October 2009 9 October 2009
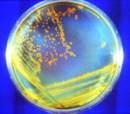
These bacteria could survive on another planet.
 D. rad Bacteria: Candidate Astronauts, photo by Michael Daly, Astronomy Picture of the Day, 30 Aug 2009. D. rad Bacteria: Candidate Astronauts, photo by Michael Daly, Astronomy Picture of the Day, 30 Aug 2009.
 Bacteria: The Space Colonists is a related local webpage. Bacteria: The Space Colonists is a related local webpage.
 9 October 2009 9 October 2009
 An asteroid covered with frozen water and organic compounds has been seen by astronomers using NASA's Infrared Telescope Facility in Hawaii. Ice on the surface should ablate away; could there be a reservoir below the surface? Could the organics be remnants of life?
An asteroid covered with frozen water and organic compounds has been seen by astronomers using NASA's Infrared Telescope Facility in Hawaii. Ice on the surface should ablate away; could there be a reservoir below the surface? Could the organics be remnants of life?
 Ice confirmed on an asteroid, by Ron Cowen, ScienceNews, 8 Oct 2009. Ice confirmed on an asteroid, by Ron Cowen, ScienceNews, 8 Oct 2009.
 More Water 'Out There:' Ice Found on Asteroid, by Nancy Atkinson, Universe Today, 8 Oct 2009. More Water 'Out There:' Ice Found on Asteroid, by Nancy Atkinson, Universe Today, 8 Oct 2009.
 Comets: The Delivery System is a related local webpage. Comets: The Delivery System is a related local webpage.
 Thanks for this alert and many others, Larry Klaes (pictured!) Thanks for this alert and many others, Larry Klaes (pictured!)
 6 October 2009 6 October 2009
Today, we know that horizontal gene transfer is a powerful evolutionary force in the microbial world, well-documented in the phylogenetic record, and one whose ecological significance is only beginning to be fully understood.... The power of horizontal gene transfer is so great that it is a major puzzle to understand why it would be that the eukaryotic world would turn its back on such a wonderful source of genetic novelty and innovation. The exciting answer, bursting through decades of dogmatic prejudice, is that it hasn't. There are now compelling documentations of horizontal gene transfer in eukaryotes, not only in plants, protists, and fungi, but in animals (including mammals) as well. The evolutionary implications have not yet been worked out, but we are confident that a fully worked out theory of the evolutionary process is required in order to properly meet the challenges posed initially by microbiology. — Microbiologists/physicists Carl Woese and Nigel Goldenfeld
When horizontal gene transfer (HGT) was first observed, it came as a surprise for darwinism. Today however, it is clear that HGT is the primary driver of evolutionary innovation among microbes. Mainstream darwinists have simply ignored the importance of this paradigm shift. Now a parallel story is now unfolding with respect to evolutionary innovation among multicelled eukaryotes — examples of HGT are accumulating, while mainstream darwinists pretend that they are rare and insignificant.
In cosmic ancestry, evolutionary innovations are possible only if the genetic programs behind them are installed already, having been acquired by some form of HGT. This acquisition may happen in steps that occur immediately before and/or long before the innovations are first expressed. Standard darwinian mutation and natural selection would affect this expression, but they would not be able to compose the programs. We think the evidence favors this theory.

 Carl R. Woese, and Nigel Goldenfeld, "How the Microbial World Saved Evolution from the Scylla of Molecular Biology and the Charybdis of the Modern Synthesis" [abstract], doi:10.1128/MMBR.00002-09, p14-21 v73, Microbiol. Mol. Biol. Rev., Mar 2009. Carl R. Woese, and Nigel Goldenfeld, "How the Microbial World Saved Evolution from the Scylla of Molecular Biology and the Charybdis of the Modern Synthesis" [abstract], doi:10.1128/MMBR.00002-09, p14-21 v73, Microbiol. Mol. Biol. Rev., Mar 2009.
 Among bacteria, HGT is "all there is" said Ernst Mayr, interviewed 7 Jun (reported 12 Jun) 2000. Among bacteria, HGT is "all there is" said Ernst Mayr, interviewed 7 Jun (reported 12 Jun) 2000.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage. Viruses and Other Gene Transfer Mechanisms is the main related local webpage.
What'sNEW about HGT  | |
 Thanks, NS, for alerting us to this paper. Thanks, NS, for alerting us to this paper.
|

 The giant [Marseillevirus, pictured] adopts genes from the other organisms, including eukaryotes, bacteria and other viruses.... — Boyer, Yutin et al.
The giant [Marseillevirus, pictured] adopts genes from the other organisms, including eukaryotes, bacteria and other viruses.... — Boyer, Yutin et al.

 In recent years, the idea has grown that horizontal transfer is the dominant force in expanding the gene repertoires of bacterial genomes. In fact, an analysis of gene phylogenies in γ-proteobacteria suggests that only a small percentage of the genes found in extant species descend from the most recent common ancestor of this group. This idea implies that microbial genomes are highly dynamic entities, constantly acquiring and losing genes. Thus... for most organisms, the percentage of genes transferred at any point in the species' history could be close to 100%, independent of size. — Otto X. Cordero and Paulien Hogeweg, Theoretical Biology and Bioinformatics, University of Utrecht, The Netherlands.
In recent years, the idea has grown that horizontal transfer is the dominant force in expanding the gene repertoires of bacterial genomes. In fact, an analysis of gene phylogenies in γ-proteobacteria suggests that only a small percentage of the genes found in extant species descend from the most recent common ancestor of this group. This idea implies that microbial genomes are highly dynamic entities, constantly acquiring and losing genes. Thus... for most organisms, the percentage of genes transferred at any point in the species' history could be close to 100%, independent of size. — Otto X. Cordero and Paulien Hogeweg, Theoretical Biology and Bioinformatics, University of Utrecht, The Netherlands. Life may make the methane on Mars. That possibility was strengthened when one possible nonbiological source, ablation from meteorites, was ruled out at Imperial College London. Two specialists in astromaterials there simulated the meteoritic process and found that it could produce only a tiny fraction of the observed methane. Other nonbiolgical sources remain under consideration.
Life may make the methane on Mars. That possibility was strengthened when one possible nonbiological source, ablation from meteorites, was ruled out at Imperial College London. Two specialists in astromaterials there simulated the meteoritic process and found that it could produce only a tiny fraction of the observed methane. Other nonbiolgical sources remain under consideration. Now the NASA team has used new tools to model the principal non-biological scenarios; they find them untenable. "We conclude that the vast majority of the nanocrystal magnetites present in the carbonate disks [example, right] could not have formed by any of the currently proposed thermal decomposition scenarios." Therefore they believe, Ancient life remains the most plausible explanation for the materials and structures found in the Mars meteorite.
Now the NASA team has used new tools to model the principal non-biological scenarios; they find them untenable. "We conclude that the vast majority of the nanocrystal magnetites present in the carbonate disks [example, right] could not have formed by any of the currently proposed thermal decomposition scenarios." Therefore they believe, Ancient life remains the most plausible explanation for the materials and structures found in the Mars meteorite. Mycoplasma pneumoniae is among the smallest bacteria that can be cultivated in vitro. Its circular genome (illustrated) measures 816,394 nucleotide pairs long, only a fifth as large as the genome of well-studied E. coli. A minimal free-living cell, M. pneumoniae was chosen by a partnership between the European Molecular Biology Laboratory in Heidelberg, Germany, and the Centro de Regulación Genómica in Barcelona, Spain, as the subject of extensive, quantitative studies. Three papers in Science report the results of these studies. One thing is clear: "Even the simplest of cells is more complex than expected."
Mycoplasma pneumoniae is among the smallest bacteria that can be cultivated in vitro. Its circular genome (illustrated) measures 816,394 nucleotide pairs long, only a fifth as large as the genome of well-studied E. coli. A minimal free-living cell, M. pneumoniae was chosen by a partnership between the European Molecular Biology Laboratory in Heidelberg, Germany, and the Centro de Regulación Genómica in Barcelona, Spain, as the subject of extensive, quantitative studies. Three papers in Science report the results of these studies. One thing is clear: "Even the simplest of cells is more complex than expected."

 The deep sea is teeming with species that have never known sunlight, according to a news release from the Census of Marine Life, a systematic investigation of life on the ocean floor. The deepest ocean floor was once thought to be too barren, cold, dark and pressurized for life to flourish there. Wrong! Actually, "The abyssal fauna is so rich in species diversity and so poorly described that collecting a known species is an anomaly," says Dr. David Billett of UK's National Oceanography Centre.
The deep sea is teeming with species that have never known sunlight, according to a news release from the Census of Marine Life, a systematic investigation of life on the ocean floor. The deepest ocean floor was once thought to be too barren, cold, dark and pressurized for life to flourish there. Wrong! Actually, "The abyssal fauna is so rich in species diversity and so poorly described that collecting a known species is an anomaly," says Dr. David Billett of UK's National Oceanography Centre. Mutations and Relative fitness over 20,000 generations (except inset)
Mutations and Relative fitness over 20,000 generations (except inset)

 An asteroid covered with frozen water and organic compounds has been seen by astronomers using NASA's Infrared Telescope Facility in Hawaii. Ice on the surface should ablate away; could there be a reservoir below the surface? Could the organics be remnants of life?
An asteroid covered with frozen water and organic compounds has been seen by astronomers using NASA's Infrared Telescope Facility in Hawaii. Ice on the surface should ablate away; could there be a reservoir below the surface? Could the organics be remnants of life?