Duplication Makes A New Primate Gene?
What'sNEW about Gene Duplication since 2005
A new primate gene has been analyzed by German molecular biologists (1). Comparing sequences from the human, mouse, rat, roundworm, fruitfly, mosquito and pufferfish genomes, the biologists identified 22 genes that were present as only single-copy orthologs in all but humans, where they have at least one paralog. These genes were most likely duplicated after the evolutionary split between the human and mouse lineages, they reasoned. Among the 22 duplicated genes, they named the most interesting one RGP. The human version of it has eight family members, each predicted to encode a protein of more than 1700 amino acids (a big one). Important functions for the proteins are likely, but not precisely known.
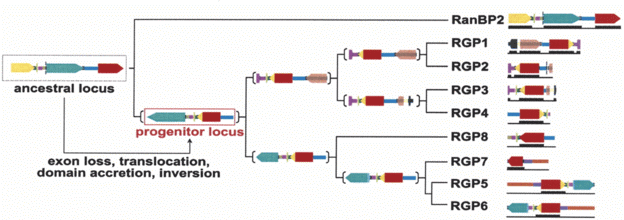 Next the biologists looked at other primates to reconstruct the more recent history of RGP. They write, "Although it is not possible to assess the temporal order of the events, the ancestor locus underwent duplication, inversion, partial deletion of the long RanBP2 exon 20, and acquisition of the 3'-end of the GCC2 gene coding for the GRIP domain.... The resulting progenitor locus contains the core coding and noncoding regions common to all the duplicated copies." At least some of the later duplications apparently occurred before the evolutionary split between the human and chimp lineages. Furthermore, "positive selection is acting on the first 20 exons of the RGP genes." That is, nucleotides which would code for different amino acids are substituted more frequently than synonymous nucleotides. The account seems well supported, and is consistent with other recent accounts for new genes.
Next the biologists looked at other primates to reconstruct the more recent history of RGP. They write, "Although it is not possible to assess the temporal order of the events, the ancestor locus underwent duplication, inversion, partial deletion of the long RanBP2 exon 20, and acquisition of the 3'-end of the GCC2 gene coding for the GRIP domain.... The resulting progenitor locus contains the core coding and noncoding regions common to all the duplicated copies." At least some of the later duplications apparently occurred before the evolutionary split between the human and chimp lineages. Furthermore, "positive selection is acting on the first 20 exons of the RGP genes." That is, nucleotides which would code for different amino acids are substituted more frequently than synonymous nucleotides. The account seems well supported, and is consistent with other recent accounts for new genes.
We would like to ask how this reconstruction fits with the mainstream darwinian theory of evolution and with our amended version, cosmic ancestry. In the latter, new genetic programs must first be installed by gene transfer. That source looks likely here, because the main progenitor gene, RANBP2, has no obvious ancestors. Rather, it is entirely absent in earlier-diverging eukaryotes like plants and fungi. Next, imported programs must be pieced together by a sophisticated software management system, as this story seems to illustrate. And the programs must then be optimized, as in the "positive selection" observed here. This process would also depend on sophisticated software management that can recognize an installed program. If the components of a genetic program may serve in more than one function, as in this example, we are not surprised. In sum, the processes described above may have merely reassembled and optimized a genetic program that already existed.
For the strictly darwinian interpretation of the history of RGP, we have questions. When point mutations were thought to be the primary source for new programs, it was possible to at least imagine stepwise paths, guided by natural selection, leading to new functions. But if wholesale recombinations of exon- and larger-sized sequences can write programs that never existed before, the first steps must be entirely hit-or-miss. How does the process find the right recombinations? How large is the sequence space of all possible recombinations? How many trials would be required? We are suspicious of the mathematics behind blind recombinations in a strictly darwinian world.
Next, when it is time for optimization within a narrower range, we do not doubt the efficacy of natural selection acting on genes that have undergone "directed mutation." This seems to be the engine behind "positive selection." But without a powerful software management system, how is the genome able to preferentially substitute for the adaptive nucletides, and not the synonymous or essential ones? Under the darwinian philosophy, this phenomenon seems teleological.
For a question we have asked before, can recombination, mutation and darwinian natural selection write new programs virtually without limit, as the history of life on Earth would seem to require? If so, closed system experiments in biology or computer models should be able to demonstrate that capability. To date, those experiments encounter a very near limit. We think those experimental results may reveal a basic fact about our world. But if the experimental environment is open to new programs supplied from elsewhere, obviously the original inventory of programs can be expanded as long as the external supply lasts.
In conclusion, the evidence concerning this new primate gene does not demonstrate that darwinian evolution can produce new genetic programs. Rather, it may demonstrate how pre-existing programs are acquired, assembled and optimized.
What'sNEW since Feb 2005
 15 May 2023: Concerted Evolution uses gene conversion to explore human duplicated genes. Does this make optimization, or even subfunctionalization, go faster? 15 May 2023: Concerted Evolution uses gene conversion to explore human duplicated genes. Does this make optimization, or even subfunctionalization, go faster?
 Extensive non-redundancy in a recently duplicated developmental gene family by E. A. Baker et al., doi:10.1186/s12862-020-01735-z, BMC Ecol Evo, 01 Mar 2021. Extensive non-redundancy in a recently duplicated developmental gene family by E. A. Baker et al., doi:10.1186/s12862-020-01735-z, BMC Ecol Evo, 01 Mar 2021.
 Duplication and divergence of the retrovirus restriction gene Fv1 in Mus caroli allows protection from multiple retroviruses by Melvyn W. Yap, George R. Young et al., doi:10.1371/journal.pgen.1008471, PLoS Genet, 11 Jun 2020. Duplication and divergence of the retrovirus restriction gene Fv1 in Mus caroli allows protection from multiple retroviruses by Melvyn W. Yap, George R. Young et al., doi:10.1371/journal.pgen.1008471, PLoS Genet, 11 Jun 2020.
 21 May 2020: Our hemoglobin evolved by duplication, divergence and recombinations.... 21 May 2020: Our hemoglobin evolved by duplication, divergence and recombinations....
 20 May 2020: Rattlesnake venom genes exemplify neofunctionalization? PNAS. 20 May 2020: Rattlesnake venom genes exemplify neofunctionalization? PNAS.
 Rapid Evolution of Gained Essential Developmental Functions of a Young Gene via Interactions with Other Essential Genes by Yuh Chwen G Lee et al., doi:10.1093/molbev/msz137, Molecular Biology and Evolution, 10 Oct 2019. Every gene in an organism's genome must have arisen at some time point in the past. ...Our study provides a novel view for how duplicated new genes can quickly become essential for viability.
Rapid Evolution of Gained Essential Developmental Functions of a Young Gene via Interactions with Other Essential Genes by Yuh Chwen G Lee et al., doi:10.1093/molbev/msz137, Molecular Biology and Evolution, 10 Oct 2019. Every gene in an organism's genome must have arisen at some time point in the past. ...Our study provides a novel view for how duplicated new genes can quickly become essential for viability.
 Molecular Dependency Impacts on the Compensating Ability of Paralogs, by Christian R. Landry and Guillaume Diss, doi:10.1016/j.tig.2017.07.012, Trends in Genetics, online 05 Sep 2017. Molecular Dependency Impacts on the Compensating Ability of Paralogs, by Christian R. Landry and Guillaume Diss, doi:10.1016/j.tig.2017.07.012, Trends in Genetics, online 05 Sep 2017.
 The evolution of duplicate gene expression in mammalian organs, by Katerina Guschanski, Maria Warnefors and Henrik Kaessmann, doi:10.1101/gr.215566.116, Genome Res., online 25 Jul 2017. The evolution of duplicate gene expression in mammalian organs, by Katerina Guschanski, Maria Warnefors and Henrik Kaessmann, doi:10.1101/gr.215566.116, Genome Res., online 25 Jul 2017.
 Gene Duplicates: Agents of Robustness or Fragility?, by R. A. Veitia, doi:10.1016/j.tig.2017.03.006, Cell, Jun 2017. Gene Duplicates: Agents of Robustness or Fragility?, by R. A. Veitia, doi:10.1016/j.tig.2017.03.006, Cell, Jun 2017.
 LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans, by Shengjun Tan et al., doi:10.1101/gr.204925.116, Genome Res, online 20 Oct 2016. Overall, our data show that LTR-mediated retroposition is highly conserved across a wide range of animal taxa.... LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans, by Shengjun Tan et al., doi:10.1101/gr.204925.116, Genome Res, online 20 Oct 2016. Overall, our data show that LTR-mediated retroposition is highly conserved across a wide range of animal taxa....
 The Phenotypic Plasticity of Duplicated Genes in Saccharomyces cerevisiae and the Origin of Adaptations, by Florian Mattenberger et al., doi:10.1534/g3.116.035329, G3, online 31 Oct 2016. The Phenotypic Plasticity of Duplicated Genes in Saccharomyces cerevisiae and the Origin of Adaptations, by Florian Mattenberger et al., doi:10.1534/g3.116.035329, G3, online 31 Oct 2016.
 23 Jul 2016: Quotation from Homology, Genes, and Evolutionary Innovation (2014), by Günter P. Wagner. 23 Jul 2016: Quotation from Homology, Genes, and Evolutionary Innovation (2014), by Günter P. Wagner.
 The Evolutionary Fates of a Large Segmental Duplication in Mouse, by Andrew P. Morgan et al., doi:10.1534/genetics.116.191007, Genetics, online 02 Jul 2016. "De novo assembly of both the ancestral (R2d1) and the derived (R2d2) copies reveals that they have been subject to non-allelic gene conversion events spanning tens of kilobases. R2d2 is also a hotspot for structural variation...." The Evolutionary Fates of a Large Segmental Duplication in Mouse, by Andrew P. Morgan et al., doi:10.1534/genetics.116.191007, Genetics, online 02 Jul 2016. "De novo assembly of both the ancestral (R2d1) and the derived (R2d2) copies reveals that they have been subject to non-allelic gene conversion events spanning tens of kilobases. R2d2 is also a hotspot for structural variation...."
 The life history of retrocopies illuminates the evolution of new mammalian genes, by Francesco Nicola Carelli et al., doi:10.1101/gr.198473.115, Genome Res., 04 Jan 2016. "Here we ...unveil the processes underlying the evolution of stripped-down retrocopies into complex new genes." The life history of retrocopies illuminates the evolution of new mammalian genes, by Francesco Nicola Carelli et al., doi:10.1101/gr.198473.115, Genome Res., 04 Jan 2016. "Here we ...unveil the processes underlying the evolution of stripped-down retrocopies into complex new genes."
 Marco Mariotti et al., "Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization" [abstract], doi:10.1101/gr.190538.115, Genome Res., 20 Jul 2015. "We show here that SPS1 genes originated through a number of independent gene duplications from an ancestral metazoan selenoprotein SPS2 gene that most likely already carried the SPS1 function." Originated? Marco Mariotti et al., "Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization" [abstract], doi:10.1101/gr.190538.115, Genome Res., 20 Jul 2015. "We show here that SPS1 genes originated through a number of independent gene duplications from an ancestral metazoan selenoprotein SPS2 gene that most likely already carried the SPS1 function." Originated?
 Raquel Assis and Doris Bachtrog, "Rapid divergence and diversification of mammalian duplicate gene functions" [html], doi:10.1186/s12862-015-0426-x, BMC Evolutionary Biology, 15 Jul 2015. Raquel Assis and Doris Bachtrog, "Rapid divergence and diversification of mammalian duplicate gene functions" [html], doi:10.1186/s12862-015-0426-x, BMC Evolutionary Biology, 15 Jul 2015.
 Matthew J Lambert et al., "Evidence for widespread subfunctionalization of splice forms in vertebrate genomes" [abstract], doi:10.1101/gr.184473.114, Genome Res., online 19 Mar 2015. "...Our results are consistent with subfunctionalization partitioning alternatively spliced isoforms among duplicate genes...." Matthew J Lambert et al., "Evidence for widespread subfunctionalization of splice forms in vertebrate genomes" [abstract], doi:10.1101/gr.184473.114, Genome Res., online 19 Mar 2015. "...Our results are consistent with subfunctionalization partitioning alternatively spliced isoforms among duplicate genes...."
 29 Apr 2014: Where Do New Genes Come From, asks Carl Zimmer.... 29 Apr 2014: Where Do New Genes Come From, asks Carl Zimmer....
 Thomas E. Keller and Soojin V. Yi, "DNA methylation and evolution of duplicate genes" [abstract], doi:10.1073/pnas.1321420111, p 5932-5937 v 111, Proc. Natl. Acad. Sci. USA, 22 Apr 2014. "Our results indicate that epigenetic modifications are intimately involved in the regulation and maintenance of duplicate genes." Thomas E. Keller and Soojin V. Yi, "DNA methylation and evolution of duplicate genes" [abstract], doi:10.1073/pnas.1321420111, p 5932-5937 v 111, Proc. Natl. Acad. Sci. USA, 22 Apr 2014. "Our results indicate that epigenetic modifications are intimately involved in the regulation and maintenance of duplicate genes."
 9 Jan 2014: ...At a key point in great ape evolution, there was a burst in retroviral activity — Edward Hollox 9 Jan 2014: ...At a key point in great ape evolution, there was a burst in retroviral activity — Edward Hollox
 David Lagman et al., "The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications" [abstract], doi:10.1186/1471-2148-13-238, v 13 n 238, BMC Evolutionary Biology, 2 Nov 2013. David Lagman et al., "The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications" [abstract], doi:10.1186/1471-2148-13-238, v 13 n 238, BMC Evolutionary Biology, 2 Nov 2013.
 Christopher R. Baker et al., "Following Gene Duplication, Paralog Interference Constrains Transcriptional Circuit Evolution" [abstract], doi:10.1126/science.1240810, p 104-108 v 342, Science, 4 Oct 2013. Christopher R. Baker et al., "Following Gene Duplication, Paralog Interference Constrains Transcriptional Circuit Evolution" [abstract], doi:10.1126/science.1240810, p 104-108 v 342, Science, 4 Oct 2013.
 8 Jun 2013: Where is the evidence for new genetic programming by standard darwinian trial-and-error? 8 Jun 2013: Where is the evidence for new genetic programming by standard darwinian trial-and-error?
 Shane Woods et al., "Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses" [html], doi:10.1371/journal.pgen.1003330, 9(5): e1003330, PLoS Genet, 30 May 2013. Shane Woods et al., "Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses" [html], doi:10.1371/journal.pgen.1003330, 9(5): e1003330, PLoS Genet, 30 May 2013.
 Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, et al., "Reconstruction of Ancestral Metabolic Enzymes Reveals Molecular Mechanisms Underlying Evolutionary Innovation through Gene Duplication" [html], doi:10.1371/journal.pbio.1001446, 10(12): e1001446, PLoS Biol., 11 Dec 2012; and commentary: Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, et al., "Reconstruction of Ancestral Metabolic Enzymes Reveals Molecular Mechanisms Underlying Evolutionary Innovation through Gene Duplication" [html], doi:10.1371/journal.pbio.1001446, 10(12): e1001446, PLoS Biol., 11 Dec 2012; and commentary:
 Richard Robinson, "Resurrecting an Ancient Enzyme to Address Gene Duplication" [html], doi:10.1371/journal.pbio.1001447, 10(12): e1001447, PLoS Biol., 11 Dec 2012. "The authors found that the ancestral maltose could split both of them, but worked much more efficiently on maltose." Richard Robinson, "Resurrecting an Ancient Enzyme to Address Gene Duplication" [html], doi:10.1371/journal.pbio.1001447, 10(12): e1001447, PLoS Biol., 11 Dec 2012. "The authors found that the ancestral maltose could split both of them, but worked much more efficiently on maltose."
 23 Oct 2012: Evolution by subfunctionalization 23 Oct 2012: Evolution by subfunctionalization
Although this model fits well with what is generally known about duplication mechanisms, it has two major limitations. First, it is difficult to formulate a process by which natural selection can sequester one member of a duplicate pair for evolutionary exploration while retaining the other for the maintenance (and/or improvement) of the ancestral function. Second, the divergence of a protein to a point at which it can no longer be detected by BLAST requires extensive substitutions along the entire length of the protein; this would occur only rarely, as many genes contain a functional protein domain that cannot easily be changed by mutations.
– Tautz and Domazet-Loŝo, 2011
|
 Diethard Tautz and Tomislav Domazet-Loŝo, "The evolutionary origin of orphan genes" [html], doi:10.1038/nrg3053, p 692-702 v 12, Nature Reviews Genetics, Oct 2011. Diethard Tautz and Tomislav Domazet-Loŝo, "The evolutionary origin of orphan genes" [html], doi:10.1038/nrg3053, p 692-702 v 12, Nature Reviews Genetics, Oct 2011.
 1 May 2010: A dual-function gene is very old according to a pair of German microbiologists. 1 May 2010: A dual-function gene is very old according to a pair of German microbiologists.
 Three New Human Genes is a related new CA webpage, posted 4 Sep 2009. Three New Human Genes is a related new CA webpage, posted 4 Sep 2009.
 Evolution's Little Helper: Xeroxed Genes, by Elizabeth Pennisi, ScienceNOW Daily News, 3 Sep 2009. Evolution's Little Helper: Xeroxed Genes, by Elizabeth Pennisi, ScienceNOW Daily News, 3 Sep 2009.
 Misook Ha et al., "Duplicate genes increase expression diversity in closely related species and allopolyploids" [abstract], doi:10.1073/pnas.0807350106, Proc. Natl. Acad. Sci. USA, online 23 Jan 2009. Misook Ha et al., "Duplicate genes increase expression diversity in closely related species and allopolyploids" [abstract], doi:10.1073/pnas.0807350106, Proc. Natl. Acad. Sci. USA, online 23 Jan 2009.
 Gavin C. Conant and Kenneth H. Wolfe, "...How duplicated genes find new functions" [abstract], doi:10.1038/nrg2482, p 938-950 v 9, Nature Reviews Genetics, Dec 2008. "Interestingly, in many cases the 'new' function of one copy is a secondary property that was always present, but that has been co-opted to a primary role after the duplication." Gavin C. Conant and Kenneth H. Wolfe, "...How duplicated genes find new functions" [abstract], doi:10.1038/nrg2482, p 938-950 v 9, Nature Reviews Genetics, Dec 2008. "Interestingly, in many cases the 'new' function of one copy is a secondary property that was always present, but that has been co-opted to a primary role after the duplication."
 David L. Des Marais and Mark D. Rausher, "Escape from adaptive conflict after duplication in an anthocyanin pathway gene" [abstract], doi:10.1038/nature07092, p 762-765 v 454, Nature, 7 Aug (online 25 Jun) 2008. "Although EAC resembles subfunctionalization in that it results in the partitioning of ancestral functions, ...EAC involves primarily adaptive substitutions." David L. Des Marais and Mark D. Rausher, "Escape from adaptive conflict after duplication in an anthocyanin pathway gene" [abstract], doi:10.1038/nature07092, p 762-765 v 454, Nature, 7 Aug (online 25 Jun) 2008. "Although EAC resembles subfunctionalization in that it results in the partitioning of ancestral functions, ...EAC involves primarily adaptive substitutions."
 Marie Sémon and Kenneth H. Wolfe, "Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis" [abstract], doi:10.1073/pnas.0708705105, Proc. Natl. Acad. Sci. USA, online 9 Jun 2008. "Surprisingly, we find that genes with slow rates of evolution are particularly prone to subfunctionalization...." Marie Sémon and Kenneth H. Wolfe, "Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis" [abstract], doi:10.1073/pnas.0708705105, Proc. Natl. Acad. Sci. USA, online 9 Jun 2008. "Surprisingly, we find that genes with slow rates of evolution are particularly prone to subfunctionalization...."
 Alexander DeLuna et al., "Exposing the fitness contribution of duplicated genes" [abstract], doi:10.1038/ng.123, p 676-681 v 40, Nature Genetics, online 13 Apr 2008. "These results suggest that most metabolic functions encoded by WGD genes are important today and were also important at the time of duplication." Alexander DeLuna et al., "Exposing the fitness contribution of duplicated genes" [abstract], doi:10.1038/ng.123, p 676-681 v 40, Nature Genetics, online 13 Apr 2008. "These results suggest that most metabolic functions encoded by WGD genes are important today and were also important at the time of duplication."
 Steffen Beisswanger and Wolfgang Stephan, "Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila" [abstract], 10.1073/pnas.0710892105, p 5447-5452 v 105, Proc. Natl. Acad. Sci. USA, 8 Apr (online 1 Apr) 2008. "Our results suggest that neofunctionalization has been achieved ...through the action of strong positive selection and the inactivation of gene conversion in part of the gene." Steffen Beisswanger and Wolfgang Stephan, "Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila" [abstract], 10.1073/pnas.0710892105, p 5447-5452 v 105, Proc. Natl. Acad. Sci. USA, 8 Apr (online 1 Apr) 2008. "Our results suggest that neofunctionalization has been achieved ...through the action of strong positive selection and the inactivation of gene conversion in part of the gene."
 20 Oct 2007: New genetic functions arise when selection is imposed on a minor side function of a preexisting gene. 20 Oct 2007: New genetic functions arise when selection is imposed on a minor side function of a preexisting gene.
 26 Sep 2007: The genomes of 17 species of fungi have been analysed to reconstruct gene duplication.... 26 Sep 2007: The genomes of 17 species of fungi have been analysed to reconstruct gene duplication....
 Jakob Lewin Rukov, Manuel Irimia et al., "High Qualitative and Quantitative Conservation of Alternative Splicing in Caenorhabditis elegans and Caenorhabditis briggsae" [abstract], 10.1093/molbev/msm023, p 909-917 v 24, Molecular Biology and Evolution, Apr (online 1 Feb) 2007. "...Gene duplication may be the major evolutionary mechanism for the origin of novel transcripts in these 2 species." Jakob Lewin Rukov, Manuel Irimia et al., "High Qualitative and Quantitative Conservation of Alternative Splicing in Caenorhabditis elegans and Caenorhabditis briggsae" [abstract], 10.1093/molbev/msm023, p 909-917 v 24, Molecular Biology and Evolution, Apr (online 1 Feb) 2007. "...Gene duplication may be the major evolutionary mechanism for the origin of novel transcripts in these 2 species."
 Eduard D. Akhunov et al., "Mechanisms and Rates of Birth and Death of Dispersed Duplicated Genes during the Evolution of a Multigene Family in Diploid and Tetraploid Wheats" [abstract], 10.1093/molbev/msl183, p 539-550 v 24, Molecular Biology and Evolution, Feb 2007 (online 29 Nov 2006). "Strong purifying selection acting on the ancestral gene ...was undiminished by the evolution of duplicated genes." Eduard D. Akhunov et al., "Mechanisms and Rates of Birth and Death of Dispersed Duplicated Genes during the Evolution of a Multigene Family in Diploid and Tetraploid Wheats" [abstract], 10.1093/molbev/msl183, p 539-550 v 24, Molecular Biology and Evolution, Feb 2007 (online 29 Nov 2006). "Strong purifying selection acting on the ancestral gene ...was undiminished by the evolution of duplicated genes."
 5 Dec 2006: Almost all human genes contain duplicated sequences. 5 Dec 2006: Almost all human genes contain duplicated sequences.
 Vaishali Katju and Michael Lynch, "On the Formation of Novel Genes by Duplication in the Caenorhabditis elegans Genome" [abstract], 10.1093/molbev/msj114, p 1056-1067 v 23, Molecular Biology and Evolution, May (online 24 Feb) 2006. "...More than 50% of newborn duplicates in Caenorhabditis elegans had unique exons in one or both members of a duplicate pair, indicating that many duplicates are not functionally identical to the progenitor copy at birth." Vaishali Katju and Michael Lynch, "On the Formation of Novel Genes by Duplication in the Caenorhabditis elegans Genome" [abstract], 10.1093/molbev/msj114, p 1056-1067 v 23, Molecular Biology and Evolution, May (online 24 Feb) 2006. "...More than 50% of newborn duplicates in Caenorhabditis elegans had unique exons in one or both members of a duplicate pair, indicating that many duplicates are not functionally identical to the progenitor copy at birth."
 Brad A. Chapman et al., "Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication" [abstract], doi: 10.1073/pnas.0507782103, Proc. Natl. Acad. Sci. USA, online 8 Feb 2006. "Contrary to classical predictions that duplicated genes may be relatively free to acquire unique functionality, we find ...that SNPs encode less radical amino acid changes in genes for which there exists a duplicated copy at a 'paleologous' locus than in 'singleton' genes. Preferential retention of duplicated genes encoding long complex proteins and their unexpectedly slow divergence ...suggest that a primary advantage of retaining duplicated paleologs may be the buffering of crucial functions." Brad A. Chapman et al., "Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication" [abstract], doi: 10.1073/pnas.0507782103, Proc. Natl. Acad. Sci. USA, online 8 Feb 2006. "Contrary to classical predictions that duplicated genes may be relatively free to acquire unique functionality, we find ...that SNPs encode less radical amino acid changes in genes for which there exists a duplicated copy at a 'paleologous' locus than in 'singleton' genes. Preferential retention of duplicated genes encoding long complex proteins and their unexpectedly slow divergence ...suggest that a primary advantage of retaining duplicated paleologs may be the buffering of crucial functions."
 30 Sep 2005: The chimp genome has been sequenced. At least seventeen human genes contain exons missing in chimps. 30 Sep 2005: The chimp genome has been sequenced. At least seventeen human genes contain exons missing in chimps.
 Lars Kuepfer et al., "Metabolic functions of duplicate genes in Saccharomyces cerevisiae" [abstract], doi: 10.1101/gr.3992505, p 1421-1430 v 15, Genome Research, Oct 2005. "Thus, at least for metabolism, persistence of the paralog fraction in the genome can be better explained with an array of different, often overlapping functional roles." Lars Kuepfer et al., "Metabolic functions of duplicate genes in Saccharomyces cerevisiae" [abstract], doi: 10.1101/gr.3992505, p 1421-1430 v 15, Genome Research, Oct 2005. "Thus, at least for metabolism, persistence of the paralog fraction in the genome can be better explained with an array of different, often overlapping functional roles."
 27 Jun 2005: "Gene duplication is the primary source of new genes." 27 Jun 2005: "Gene duplication is the primary source of new genes."
 28 Feb 2005: Can pre-existing genetic programs be pieced together? 28 Feb 2005: Can pre-existing genetic programs be pieced together?
 21 Feb 2005: Duplication Makes A New Primate Gene — our What'sNEW announcement of this webpage. 21 Feb 2005: Duplication Makes A New Primate Gene — our What'sNEW announcement of this webpage.
 17 Jan 2003: Duplicated genes serve backup functions. 17 Jan 2003: Duplicated genes serve backup functions.
References
1. Francesca D. Ciccarelli et al., "Complex genomic rearrangements lead to novel primate gene function" [abstract], DOI: 10.1101/gr.3266405, Genome Research, online 14 Feb 2005. Also see: Scientists Document Complex Genomic Events Leading To The Birth Of New Genes, ScienceDaily, 15 Feb 2005.
![]()
|
 Next the biologists looked at other primates to reconstruct the more recent history of RGP. They write, "Although it is not possible to assess the temporal order of the events, the ancestor locus underwent duplication, inversion, partial deletion of the long RanBP2 exon 20, and acquisition of the 3'-end of the GCC2 gene coding for the GRIP domain.... The resulting progenitor locus contains the core coding and noncoding regions common to all the duplicated copies." At least some of the later duplications apparently occurred before the evolutionary split between the human and chimp lineages. Furthermore, "positive selection is acting on the first 20 exons of the RGP genes." That is, nucleotides which would code for different amino acids are substituted more frequently than synonymous nucleotides. The account seems well supported, and is consistent with other recent accounts for new genes.
Next the biologists looked at other primates to reconstruct the more recent history of RGP. They write, "Although it is not possible to assess the temporal order of the events, the ancestor locus underwent duplication, inversion, partial deletion of the long RanBP2 exon 20, and acquisition of the 3'-end of the GCC2 gene coding for the GRIP domain.... The resulting progenitor locus contains the core coding and noncoding regions common to all the duplicated copies." At least some of the later duplications apparently occurred before the evolutionary split between the human and chimp lineages. Furthermore, "positive selection is acting on the first 20 exons of the RGP genes." That is, nucleotides which would code for different amino acids are substituted more frequently than synonymous nucleotides. The account seems well supported, and is consistent with other recent accounts for new genes.